CDBProm: the Comprehensive Directory of Bacterial Promoters
- PMID: 38385146
- PMCID: PMC10880602
- DOI: 10.1093/nargab/lqae018
CDBProm: the Comprehensive Directory of Bacterial Promoters
Abstract
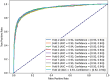
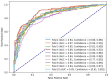
The decreasing cost of whole genome sequencing has produced high volumes of genomic information that require annotation. The experimental identification of promoter sequences, pivotal for regulating gene expression, is a laborious and cost-prohibitive task. To expedite this, we introduce the Comprehensive Directory of Bacterial Promoters (CDBProm), a directory of in-silico predicted bacterial promoter sequences. We first identified that an Extreme Gradient Boosting (XGBoost) algorithm would distinguish promoters from random downstream regions with an accuracy of 87%. To capture distinctive promoter signals, we generated a second XGBoost classifier trained on the instances misclassified in our first classifier. The predictor of CDBProm is then fed with over 55 million upstream regions from more than 6000 bacterial genomes. Upon finding potential promoter sequences in upstream regions, each promoter is mapped to the genomic data of the organism, linking the predicted promoter with its coding DNA sequence, and identifying the function of the gene regulated by the promoter. The collection of bacterial promoters available in CDBProm enables the quantitative analysis of a plethora of bacterial promoters. Our collection with over 24 million promoters is publicly available at https://aw.iimas.unam.mx/cdbprom/.
© The Author(s) 2024. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures



References
-
- Cases I., de Lorenzo V., Ouzounis C.A.. Transcription regulation and environmental adaptation in bacteria. Trends Microbiol. 2003; 11:248–253. - PubMed
-
- Barnard A., Wolfe A., Busby S.. Regulation at complex bacterial promoters: how bacteria use different promoter organizations to produce different regulatory outcomes. Curr. Opin. Microbiol. 2004; 7:102–108. - PubMed
-
- Krebs J.E., Goldstein E.S., Kilpatrick S.T.. Lewin's Gene XII. 2017; 12th ednBurlington: Jones & Bartlett Learning.
LinkOut - more resources
Full Text Sources

