High-Dimensional Single-Cell Multimodal Landscape of Human Carotid Atherosclerosis
- PMID: 38385291
- PMCID: PMC10978277
- DOI: 10.1161/ATVBAHA.123.320524
High-Dimensional Single-Cell Multimodal Landscape of Human Carotid Atherosclerosis
Abstract
Background: Atherosclerotic plaques are complex tissues composed of a heterogeneous mixture of cells. However, our understanding of the comprehensive transcriptional and phenotypic landscape of the cells within these lesions is limited.
Methods: To characterize the landscape of human carotid atherosclerosis in greater detail, we combined cellular indexing of transcriptomes and epitopes by sequencing and single-cell RNA sequencing to classify all cell types within lesions (n=21; 13 symptomatic) to achieve a comprehensive multimodal understanding of the cellular identities of atherosclerosis and their association with clinical pathophysiology.
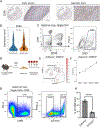
Results: We identified 25 cell populations, each with a unique multiomic signature, including macrophages, T cells, NK (natural killer) cells, mast cells, B cells, plasma cells, neutrophils, dendritic cells, endothelial cells, fibroblasts, and smooth muscle cells (SMCs). Among the macrophages, we identified 2 proinflammatory subsets enriched in IL-1B (interleukin-1B) or C1Q expression, 2 TREM2-positive foam cells (1 expressing inflammatory genes), and subpopulations with a proliferative gene signature and SMC-specific gene signature with fibrotic pathways upregulated. Further characterization revealed various subsets of SMCs and fibroblasts, including SMC-derived foam cells. These foamy SMCs were localized in the deep intima of coronary atherosclerotic lesions. Utilizing cellular indexing of transcriptomes and epitopes by sequencing data, we developed a flow cytometry panel, using cell surface proteins CD29, CD142, and CD90, to isolate SMC-derived cells from lesions. Lastly, we observed reduced proportions of efferocytotic macrophages, classically activated endothelial cells, and contractile and modulated SMC-derived cells, while inflammatory SMCs were enriched in plaques of clinically symptomatic versus asymptomatic patients.
Conclusions: Our multimodal atlas of cell populations within atherosclerosis provides novel insights into the diversity, phenotype, location, isolation, and clinical relevance of the unique cellular composition of human carotid atherosclerosis. These findings facilitate both the mapping of cardiovascular disease susceptibility loci to specific cell types and the identification of novel molecular and cellular therapeutic targets for the treatment of the disease.
Keywords: atherosclerosis; fibroblasts; macrophage; single-cell analysis; smooth muscle cell.
Conflict of interest statement
Figures





Update of
-
High-Dimensional Single-Cell Multimodal Landscape of Human Carotid Atherosclerosis.medRxiv [Preprint]. 2023 Jul 16:2023.07.13.23292633. doi: 10.1101/2023.07.13.23292633. medRxiv. 2023. Update in: Arterioscler Thromb Vasc Biol. 2024 Apr;44(4):930-945. doi: 10.1161/ATVBAHA.123.320524. PMID: 37502836 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

