Pathogenic and genomic characterization of rabbit-sourced Pasteurella multocida serogroup F isolates recovered from dead rabbits with respiratory disease
- PMID: 38385714
- PMCID: PMC10986509
- DOI: 10.1128/spectrum.03654-23
Pathogenic and genomic characterization of rabbit-sourced Pasteurella multocida serogroup F isolates recovered from dead rabbits with respiratory disease
Abstract
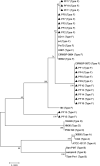
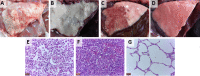
Pasteurella multocida serogroup F can infect a number of animals. However, the pathogenicity and genomic features of this serogroup are still largely unknown. In the present study, the pathogenicity and genomic sequences of 19 rabbit-sourced P. multocida serogroup F isolates were determined. The 19 isolates were highly pathogenic for rabbits causing severe pathologic lesions and high mortality in inoculated rabbits. Nevertheless, the pathologic lesions in rabbits caused by the 19 isolates were distinct from those caused by the previously reported high-virulent serogroup F strains J-4103 (rabbit), P-4218 (turkey), and C21724H3km7 (chicken). Moreover, the 19 isolates were avirulent to white feather broilers. The genomes of the 19 isolates were determined to understand the pathogenicity of these isolates. The finding of a number of functional genes in the 19 isolates by comparison with the low-virulent rabbit-sourced serogroup F strain s4 might contribute to the high virulence of these isolates. Notably, polymorphisms were determined in the lipopolysaccharide outer core biosynthetic genes natC and gatF among the serogroup F strains of different hosts. However, the sequences of natC and gatF from rabbit-sourced strains (except for SD11) were identical, which might be responsible for the host specific of the 19 isolates. The observations and findings in this study would be helpful for the understanding of the pathogenicity variation and host predilection of P. multocida.
Importance: The 19 rabbit-sourced Pasteurella multocida serogroup F isolates showing high virulence to rabbits were avirulent to the broilers. Notably, polymorphisms were determined in the lipopolysaccharide outer core biosynthetic genes natC and gatF among all serogroup F strains of different hosts. However, the sequences of natC and gatF from rabbit-sourced strains (except for SD11) were identical, which might be responsible for the host specific of the 19 isolates.
Keywords: Pasteurella multocida serogroup F; pathogenicity; rabbit; whole-genome sequence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Pathogenic and genomic characterisation of a rabbit sourced Pasteurella multocida serogroup F isolate s4.BMC Vet Res. 2022 Jul 23;18(1):288. doi: 10.1186/s12917-022-03381-7. BMC Vet Res. 2022. PMID: 35869529 Free PMC article.
-
Experimental pathogenicity and complete genome characterization of a pig origin Pasteurella multocida serogroup F isolate HN07.Vet Microbiol. 2017 Jan;198:23-33. doi: 10.1016/j.vetmic.2016.11.028. Epub 2016 Nov 27. Vet Microbiol. 2017. PMID: 28062004
-
Host response in rabbits to infection with Pasteurella multocida serogroup F strains originating from fowl cholera.Can J Vet Res. 2011 Jul;75(3):200-8. Can J Vet Res. 2011. PMID: 22210996 Free PMC article.
-
Pasteurella multocida: Genotypes and Genomics.Microbiol Mol Biol Rev. 2019 Sep 4;83(4):e00014-19. doi: 10.1128/MMBR.00014-19. Print 2019 Nov 20. Microbiol Mol Biol Rev. 2019. PMID: 31484691 Free PMC article. Review.
-
Pathogenomics insights for understanding Pasteurella multocida adaptation.Int J Med Microbiol. 2020 May;310(4):151417. doi: 10.1016/j.ijmm.2020.151417. Epub 2020 Mar 25. Int J Med Microbiol. 2020. PMID: 32276876 Review.
Cited by
-
Virulence and resistance gene analysis of Rothia nasimurium by whole gene sequencing.Sci Rep. 2025 Mar 27;15(1):10583. doi: 10.1038/s41598-025-95405-z. Sci Rep. 2025. PMID: 40148435 Free PMC article.
References
-
- Rhoades KR, Rimler RB. 1987. Capsular groups of Pasteurella multocida isolated from avian hosts. Avian Dis 31:895–898. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

