Comprehensive performance evaluation of ligand-binding assay-LC-MS/MS method for co-dosed monoclonal anti-SARS-CoV-2 antibodies (AZD7442)
- PMID: 38385904
- PMCID: PMC11845114
- DOI: 10.4155/bio-2023-0225
Comprehensive performance evaluation of ligand-binding assay-LC-MS/MS method for co-dosed monoclonal anti-SARS-CoV-2 antibodies (AZD7442)
Abstract
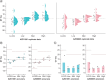
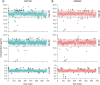
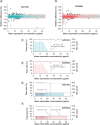
Aims: AZD7442 is a combination SARS-CoV-2 therapy comprising two co-dosed monoclonal antibodies. Materials & methods: The authors validated a hybrid ligand-binding assay-LC-MS/MS method for pharmacokinetic assessment of AZD7442 in human serum with nominal concentration range of each analyte of 0.300-30.0 μg/ml. Results: Validation results met current regulatory acceptance criteria. The validated method supported three clinical trials that spanned more than 17 months and ≥720 analytical runs (∼30,000 samples and ∼3000 incurred sample reanalyses per analyte). The data generated supported multiple health authority interactions, across the globe. AZD7442 (EVUSHELD) was approved in 12 countries for pre-exposure prophylaxis of COVID-19. Conclusion: The results reported here demonstrate the robust, high-throughput capability of the hybrid ligand-binding assay-LC-MS/MS approach being employed to support-next generation versions of EVUSHELD, AZD3152.
Keywords: SARS-CoV-2; hybrid IA–LC–MS/MS; hybrid LBA–LC–MS/MS; incurred sample reanalysis; monoclonal antibody; pharmacokinetics; tixagevimab and cilgavimab; validation.
Plain language summary
The measurement of antibodies in human body fluids (e.g., blood, serum) has historically been tied to laboratory tests that may face operational limitations, including susceptibility to interference from other blood components and a reliance on unique reagents that can take months to produce. As such, there is a pursuit of alternative analytical methods to more accurately detect and measure antibody drugs from complex matrices. In the method, the authors describe different techniques that once combined were used to capture, separate, filter, fragment and then detect and measure the co-dosed antibody drugs. This method has been validated in accordance with current health authority guidelines and has been used to support three clinical trials that spanned more than 17 months; that is, the validated method was used to analyze nearly 30,000 serum samples from more than 2000 patients. Collectively, the results reported here demonstrate the robustness and high-throughput capability of this analytical approach.
Conflict of interest statement
Authors Y Huang, CC Wang and AI Rosenbaum performed work within this manuscript while employed by AstraZeneca. The authors’ employment and stock investments in AstraZeneca do not affect the authenticity and objectivity of the experimental result detailed in this manuscript. Authors MS Woolf, SM Naser, AM Wheeler, WR Mylott and E Ma performed work within this manuscript while employed by PPD, a part of Thermo Fisher Scientific. The authors’ employment and stock investments in Thermo Fisher Scientific do not affect the authenticity and objectivity of the experimental result detailed in this manuscript. Responsibility for opinions, conclusions and data interpretation lies with the authors and should not be interpreted as representing the official views or policies of the Department of Defense or the US government. The authors have no other competing interests or relevant affiliations with any organization or entity with the subject matter or materials discussed in the manuscript apart from those disclosed.
Figures



Similar articles
-
Multiplex Hybrid Antigen-Capture LC-MRM Quantification in Sera and Nasal Lining Fluid of AZD7442, a SARS-CoV-2-Targeting Antibody Combination.Anal Chem. 2022 Nov 1;94(43):14835-14845. doi: 10.1021/acs.analchem.2c01320. Epub 2022 Oct 21. Anal Chem. 2022. PMID: 36269894 Free PMC article.
-
Safety, Tolerability and Pharmacokinetics of Half-Life Extended Severe Acute Respiratory Syndrome Coronavirus 2 Neutralizing Monoclonal Antibodies AZD7442 (Tixagevimab-Cilgavimab) in Healthy Adults.J Infect Dis. 2023 May 12;227(10):1153-1163. doi: 10.1093/infdis/jiad014. J Infect Dis. 2023. PMID: 36683419 Free PMC article. Clinical Trial.
-
Qualification of a Biolayer Interferometry Assay to Support AZD7442 Resistance Monitoring.Microbiol Spectr. 2022 Oct 26;10(5):e0103422. doi: 10.1128/spectrum.01034-22. Epub 2022 Aug 22. Microbiol Spectr. 2022. PMID: 35993765 Free PMC article.
-
A Critical Analysis of the Use of Cilgavimab plus Tixagevimab Monoclonal Antibody Cocktail (Evusheld™) for COVID-19 Prophylaxis and Treatment.Viruses. 2022 Sep 9;14(9):1999. doi: 10.3390/v14091999. Viruses. 2022. PMID: 36146805 Free PMC article. Review.
-
The efficacy of tixagevimab/cilgavimab (Evusheld) in prophylaxis and treatment of COVID-19 in immunocompromised patients: a systematic review and meta-analysis.Eur J Med Res. 2024 Jan 5;29(1):27. doi: 10.1186/s40001-023-01549-x. Eur J Med Res. 2024. PMID: 38183123 Free PMC article.
References
-
- Mullard A. FDA approves 100th monoclonal antibody product. Nat. Rev. Drug Discov. 20(7), 491–495 (2021). - PubMed
-
• This retrospective article describes the emergence and advancement of therapeutic monoclonal antibody products.
-
- Todd PA , Brogden RN. Muromonab CD3. Drugs 37(6), 871–899 (1989). - PubMed
-
- Singh S , Tank NK , Dwiwedi Pet al. . Monoclonal antibodies: a review. Curr. Clin. Pharmacol. 13(2), 85–99 (2018). - PubMed
-
- Lindsley CW. Predictions and statistics for the best-selling drugs globally and in the United States in 2018 and a look forward to 2024 projections. ACS Chemical Neuroscience 10(3), 1115–1115 (2019). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
