Unlocking allelic variation in circadian clock genes to develop environmentally robust and productive crops
- PMID: 38386103
- PMCID: PMC10884192
- DOI: 10.1007/s00425-023-04324-8
Unlocking allelic variation in circadian clock genes to develop environmentally robust and productive crops
Abstract
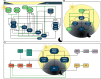
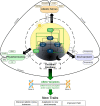
Molecular mechanisms of biological rhythms provide opportunities to harness functional allelic diversity in core (and trait- or stress-responsive) oscillator networks to develop more climate-resilient and productive germplasm. The circadian clock senses light and temperature in day-night cycles to drive biological rhythms. The clock integrates endogenous signals and exogenous stimuli to coordinate diverse physiological processes. Advances in high-throughput non-invasive assays, use of forward- and inverse-genetic approaches, and powerful algorithms are allowing quantitation of variation and detection of genes associated with circadian dynamics. Circadian rhythms and phytohormone pathways in response to endogenous and exogenous cues have been well documented the model plant Arabidopsis. Novel allelic variation associated with circadian rhythms facilitates adaptation and range expansion, and may provide additional opportunity to tailor climate-resilient crops. The circadian phase and period can determine adaptation to environments, while the robustness in the circadian amplitude can enhance resilience to environmental changes. Circadian rhythms in plants are tightly controlled by multiple and interlocked transcriptional-translational feedback loops involving morning (CCA1, LHY), mid-day (PRR9, PRR7, PRR5), and evening (TOC1, ELF3, ELF4, LUX) genes that maintain the plant circadian clock ticking. Significant progress has been made to unravel the functions of circadian rhythms and clock genes that regulate traits, via interaction with phytohormones and trait-responsive genes, in diverse crops. Altered circadian rhythms and clock genes may contribute to hybrid vigor as shown in Arabidopsis, maize, and rice. Modifying circadian rhythms via transgenesis or genome-editing may provide additional opportunities to develop crops with better buffering capacity to environmental stresses. Models that involve clock gene‒phytohormone‒trait interactions can provide novel insights to orchestrate circadian rhythms and modulate clock genes to facilitate breeding of all season crops.
Keywords: Adaptation; Allelic variation; Biological rhythms; Breeding all season crops; Clock genes and signaling; Heterosis; Non-invasive assays; Stress tolerance.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests or conflicts of interest, relating to funding, employment or financial (or non-financial) interests. The authors have no relevant financial or non-financial interests to disclose. The authors have no competing interests to declare that are relevant to the content of this article. All authors certify that they have no affiliations with or involvement in any organization or entity with any financial interest or non-financial interest in the subject matter or materials discussed in this manuscript. The authors have no financial or proprietary interests in any material discussed in this article.
Figures


Similar articles
-
Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis.Curr Biol. 2011 Jan 25;21(2):120-5. doi: 10.1016/j.cub.2010.12.013. Epub 2011 Jan 13. Curr Biol. 2011. PMID: 21236675 Free PMC article.
-
A genetic study of the Arabidopsis circadian clock with reference to the TIMING OF CAB EXPRESSION 1 (TOC1) gene.Plant Cell Physiol. 2009 Feb;50(2):290-303. doi: 10.1093/pcp/pcn198. Epub 2008 Dec 19. Plant Cell Physiol. 2009. PMID: 19098071
-
REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock.PLoS Genet. 2011 Mar;7(3):e1001350. doi: 10.1371/journal.pgen.1001350. Epub 2011 Mar 31. PLoS Genet. 2011. PMID: 21483796 Free PMC article.
-
The Roles of Circadian Clock Genes in Plant Temperature Stress Responses.Int J Mol Sci. 2024 Jan 11;25(2):918. doi: 10.3390/ijms25020918. Int J Mol Sci. 2024. PMID: 38255990 Free PMC article. Review.
-
Circadian Rhythms in Plants.Cold Spring Harb Perspect Biol. 2019 Sep 3;11(9):a034611. doi: 10.1101/cshperspect.a034611. Cold Spring Harb Perspect Biol. 2019. PMID: 31138544 Free PMC article. Review.
Cited by
-
Complex Signaling Networks Underlying Blue-Light-Mediated Floral Transition in Plants.Plants (Basel). 2025 May 20;14(10):1533. doi: 10.3390/plants14101533. Plants (Basel). 2025. PMID: 40431098 Free PMC article. Review.
-
Estimating Genetic Variability and Heritability of Morpho-Agronomic Traits of M5 Cowpea (Vigna unguiculata (L.) Walp) Mutant Lines.Int J Mol Sci. 2025 Aug 5;26(15):7543. doi: 10.3390/ijms26157543. Int J Mol Sci. 2025. PMID: 40806671 Free PMC article.
-
A review on modeling approaches for the transcriptional regulatory network intricacies of circadian clock genes in plants.Planta. 2025 Jun 5;262(1):17. doi: 10.1007/s00425-025-04735-9. Planta. 2025. PMID: 40471439 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

