3D Genome Reconstruction from Partially Phased Hi-C Data
- PMID: 38386111
- PMCID: PMC10884149
- DOI: 10.1007/s11538-024-01263-7
3D Genome Reconstruction from Partially Phased Hi-C Data
Abstract
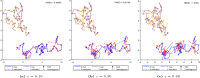
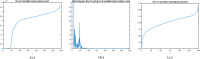
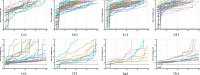
The 3-dimensional (3D) structure of the genome is of significant importance for many cellular processes. In this paper, we study the problem of reconstructing the 3D structure of chromosomes from Hi-C data of diploid organisms, which poses additional challenges compared to the better-studied haploid setting. With the help of techniques from algebraic geometry, we prove that a small amount of phased data is sufficient to ensure finite identifiability, both for noiseless and noisy data. In the light of these results, we propose a new 3D reconstruction method based on semidefinite programming, paired with numerical algebraic geometry and local optimization. The performance of this method is tested on several simulated datasets under different noise levels and with different amounts of phased data. We also apply it to a real dataset from mouse X chromosomes, and we are then able to recover previously known structural features.
Keywords: 3D genome organization; Applied algebraic geometry; Diploid organisms; Hi-C; Numerical algebraic geometry.
© 2024. The Author(s).
Figures


References
-
- Alfakih AY, Khandani A, Wolkowicz H. Solving euclidean distance matrix completion problems via semidefinite programming. Comput Optim Appl. 1999;12(1):13–30. doi: 10.1023/A:1008655427845. - DOI
-
- Belyaeva A, Kubjas K, Sun LJ, Uhler C. Identifying 3D genome organization in diploid organisms via Euclidean distance geometry. SIAM J Math Data Sci. 2022;4(1):204–228. doi: 10.1137/21M1390372. - DOI
-
- Breiding P, Rose K, Timme S. Certifying zeros of polynomial systems using interval arithmetic. ACM Trans Math Softw. 2023;49(1):1–14. doi: 10.1145/3580277. - DOI
-
- Breiding P, Timme S (2018) HomotopyContinuation.jl: A package for homotopy continuation in Julia. In: Davenport JH, Kauers M, Labahn G, Urban J (eds) Mathematical Software—ICMS 2018. Springer, Cham, pp 458–465
-
- Cauer AG, Yardimci G, Vert JP, Varoquaux N, Noble WS (2019) Inferring diploid 3D chromatin structures from Hi-C data. In: 19th International workshop on algorithms in bioinformatics (WABI 2019)

