Broad Next-Generation Integrated Sequencing of Myelofibrosis Identifies Disease-Specific and Age-Related Genomic Alterations
- PMID: 38386293
- PMCID: PMC11061602
- DOI: 10.1158/1078-0432.CCR-23-0372
Broad Next-Generation Integrated Sequencing of Myelofibrosis Identifies Disease-Specific and Age-Related Genomic Alterations
Abstract
Purpose: Myeloproliferative neoplasms (MPN) are characterized by the overproduction of differentiated myeloid cells. Mutations in JAK2, CALR, and MPL are considered drivers of Bcr-Abl-ve MPN, including essential thrombocythemia (ET), polycythemia vera (PV), prefibrotic primary myelofibrosis (prePMF), and overt myelofibrosis (MF). However, how these driver mutations lead to phenotypically distinct and/or overlapping diseases is unclear.
Experimental design: To compare the genetic landscape of MF to ET/PV/PrePMF, we sequenced 1,711 genes for mutations along with whole transcriptome RNA sequencing of 137 patients with MPN.
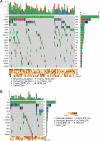
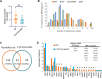
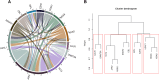
Results: In addition to driver mutations, 234 and 74 genes were found to be mutated in overt MF (N = 106) and ET/PV/PrePMF (N = 31), respectively. Overt MF had more mutations compared with ET/PV/prePMF (5 vs. 4 per subject, P = 0.006). Genes frequently mutated in MF included high-risk genes (ASXL1, SRSF2, EZH2, IDH1/2, and U2AF1) and Ras pathway genes. Mutations in NRAS, KRAS, SRSF2, EZH2, IDH2, and NF1 were exclusive to MF. Advancing age, higher DIPSS, and poor overall survival (OS) correlated with increased variants in MF. Ras mutations were associated with higher leukocytes and platelets and poor OS. The comparison of gene expression showed upregulation of proliferation and inflammatory pathways in MF. Notably, ADGRL4, DNASE1L3, PLEKHGB4, HSPG2, MAMDC2, and DPYSL3 were differentially expressed in hematopoietic stem and differentiated cells.
Conclusions: Our results illustrate that evolution of MF from ET/PV/PrePMF likely advances with age, accumulation of mutations, and activation of proliferative pathways. The genes and pathways identified by integrated genomics approach provide insight into disease transformation and progression and potential targets for therapeutic intervention.
©2024 The Authors; Published by the American Association for Cancer Research.
Figures




References
-
- Vannucchi AM, Lasho TL, Guglielmelli P, Biamonte F, Pardanani A, Pereira A, et al. . Mutations and prognosis in primary myelofibrosis. Leukemia 2013;27:1861–9. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

