ZSCAN10 deficiency causes a neurodevelopmental disorder with characteristic oto-facial malformations
- PMID: 38386308
- PMCID: PMC11224597
- DOI: 10.1093/brain/awae058
ZSCAN10 deficiency causes a neurodevelopmental disorder with characteristic oto-facial malformations
Abstract
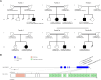
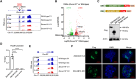
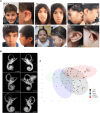
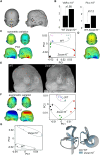
Neurodevelopmental disorders are major indications for genetic referral and have been linked to more than 1500 loci including genes encoding transcriptional regulators. The dysfunction of transcription factors often results in characteristic syndromic presentations; however, at least half of these patients lack a genetic diagnosis. The implementation of machine learning approaches has the potential to aid in the identification of new disease genes and delineate associated phenotypes. Next generation sequencing was performed in seven affected individuals with neurodevelopmental delay and dysmorphic features. Clinical characterization included reanalysis of available neuroimaging datasets and 2D portrait image analysis with GestaltMatcher. The functional consequences of ZSCAN10 loss were modelled in mouse embryonic stem cells (mESCs), including a knockout and a representative ZSCAN10 protein truncating variant. These models were characterized by gene expression and western blot analyses, chromatin immunoprecipitation and quantitative PCR (ChIP-qPCR) and immunofluorescence staining. Zscan10 knockout mouse embryos were generated and phenotyped. We prioritized bi-allelic ZSCAN10 loss-of-function variants in seven affected individuals from five unrelated families as the underlying molecular cause. RNA-sequencing analyses in Zscan10-/- mESCs indicated dysregulation of genes related to stem cell pluripotency. In addition, we established in mESCs the loss-of-function mechanism for a representative human ZSCAN10 protein truncating variant by showing alteration of its expression levels and subcellular localization, interfering with its binding to DNA enhancer targets. Deep phenotyping revealed global developmental delay, facial asymmetry and malformations of the outer ear as consistent clinical features. Cerebral MRI showed dysplasia of the semicircular canals as an anatomical correlate of sensorineural hearing loss. Facial asymmetry was confirmed as a clinical feature by GestaltMatcher and was recapitulated in the Zscan10 mouse model along with inner and outer ear malformations. Our findings provide evidence of a novel syndromic neurodevelopmental disorder caused by bi-allelic loss-of-function variants in ZSCAN10.
Keywords: neurodevelopmental disorders; oto-facial syndrome; semicircular canal dysplasia; zinc finger transcription factor.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
B.B. is Co-Founder and Shareholder of AIRAmed GmbH. The other authors report no competing interests.
Figures
References
-
- Sheridan E, Wright J, Small N, et al. Risk factors for congenital anomaly in a multiethnic birth cohort: An analysis of the Born in Bradford study. Lancet. 2013;382:1350–1359. - PubMed
-
- Maulik PK, Mascarenhas MN, Mathers CD, Dua T, Saxena S. Prevalence of intellectual disability: A meta-analysis of population-based studies. Res Dev Disabil. 2011;32:419–436. - PubMed

