Sall genes regulate hindlimb initiation in mouse embryos
- PMID: 38386912
- PMCID: PMC11075541
- DOI: 10.1093/genetics/iyae029
Sall genes regulate hindlimb initiation in mouse embryos
Abstract
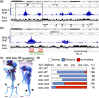
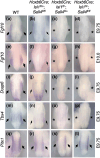
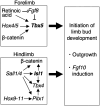
Vertebrate limbs start to develop as paired protrusions from the lateral plate mesoderm at specific locations of the body with forelimb buds developing anteriorly and hindlimb buds posteriorly. During the initiation process, limb progenitor cells maintain active proliferation to form protrusions and start to express Fgf10, which triggers molecular processes for outgrowth and patterning. Although both processes occur in both types of limbs, forelimbs (Tbx5), and hindlimbs (Isl1) utilize distinct transcriptional systems to trigger their development. Here, we report that Sall1 and Sall4, zinc finger transcription factor genes, regulate hindlimb initiation in mouse embryos. Compared to the 100% frequency loss of hindlimb buds in TCre; Isl1 conditional knockouts, Hoxb6Cre; Isl1 conditional knockout causes a hypomorphic phenotype with only approximately 5% of mutants lacking the hindlimb. Our previous study of SALL4 ChIP-seq showed SALL4 enrichment in an Isl1 enhancer, suggesting that SALL4 acts upstream of Isl1. Removing 1 allele of Sall4 from the hypomorphic Hoxb6Cre; Isl1 mutant background caused loss of hindlimbs, but removing both alleles caused an even higher frequency of loss of hindlimbs, suggesting a genetic interaction between Sall4 and Isl1. Furthermore, TCre-mediated conditional double knockouts of Sall1 and Sall4 displayed a loss of expression of hindlimb progenitor markers (Isl1, Pitx1, Tbx4) and failed to develop hindlimbs, demonstrating functional redundancy between Sall1 and Sall4. Our data provides genetic evidence that Sall1 and Sall4 act as master regulators of hindlimb initiation.
Keywords: Isl1; Sall1; Sall4; hindlimb; limb initiation.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America. All rights reserved. For commercial re-use, please contact reprints@oup.com for reprints and translation rights for reprints. All other permissions can be obtained through our RightsLink service via the Permissions link on the article page on our site—for further information please contact journals.permissions@oup.com.
Conflict of interest statement
Conflicts of interest: The authors declare no conflicts of interest.
Figures


Similar articles
-
Sall4-Gli3 system in early limb progenitors is essential for the development of limb skeletal elements.Proc Natl Acad Sci U S A. 2015 Apr 21;112(16):5075-80. doi: 10.1073/pnas.1421949112. Epub 2015 Apr 6. Proc Natl Acad Sci U S A. 2015. PMID: 25848055 Free PMC article.
-
Novel function of Hox13 in regulating outgrowth of the newt hindlimb bud through interaction with Fgf10 and Tbx4.Dev Growth Differ. 2025 Jan;67(1):10-22. doi: 10.1111/dgd.12952. Epub 2024 Dec 26. Dev Growth Differ. 2025. PMID: 39725403 Free PMC article.
-
Isl1 and Ldb co-regulators of transcription are essential early determinants of mouse limb development.Dev Dyn. 2012 Apr;241(4):787-91. doi: 10.1002/dvdy.23761. Dev Dyn. 2012. PMID: 22411555 Free PMC article.
-
How the embryo makes a limb: determination, polarity and identity.J Anat. 2015 Oct;227(4):418-30. doi: 10.1111/joa.12361. Epub 2015 Aug 7. J Anat. 2015. PMID: 26249743 Free PMC article. Review.
-
Subdivision of the lateral plate mesoderm and specification of the forelimb and hindlimb forming domains.Semin Cell Dev Biol. 2016 Jan;49:102-8. doi: 10.1016/j.semcdb.2015.11.011. Epub 2015 Nov 28. Semin Cell Dev Biol. 2016. PMID: 26643124 Review.
Cited by
-
Zebrafish sall1a and sall4 contribute to body elongation.Dev Biol. 2025 Sep;525:185-193. doi: 10.1016/j.ydbio.2025.06.006. Epub 2025 Jun 5. Dev Biol. 2025. PMID: 40482694
References
-
- Agresti A. 2012. Categorical Data Analysis. Hoboken (NJ): Wiley. p. 69–112.
-
- Chen KQ, Tahara N, Anderson A, Kawakami H, Kawakami S, Nishinakamura R, Pandolfi PP, Kawakami Y. 2020. Development of the proximal-anterior skeletal elements in the mouse hindlimb is regulated by a transcriptional and signaling network controlled by sall4. Genetics. 215(1):129–141. doi:10.1534/genetics.120.303069. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

