Semi-supervised auto-segmentation method for pelvic organ-at-risk in magnetic resonance images based on deep-learning
- PMID: 38386963
- PMCID: PMC10929999
- DOI: 10.1002/acm2.14296
Semi-supervised auto-segmentation method for pelvic organ-at-risk in magnetic resonance images based on deep-learning
Abstract
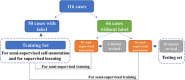
Background and purpose: In radiotherapy, magnetic resonance (MR) imaging has higher contrast for soft tissues compared to computed tomography (CT) scanning and does not emit radiation. However, manual annotation of the deep learning-based automatic organ-at-risk (OAR) delineation algorithms is expensive, making the collection of large-high-quality annotated datasets a challenge. Therefore, we proposed the low-cost semi-supervised OAR segmentation method using small pelvic MR image annotations.
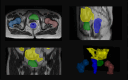
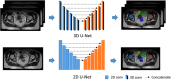
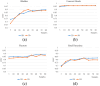
Methods: We trained a deep learning-based segmentation model using 116 sets of MR images from 116 patients. The bladder, femoral heads, rectum, and small intestine were selected as OAR regions. To generate the training set, we utilized a semi-supervised method and ensemble learning techniques. Additionally, we employed a post-processing algorithm to correct the self-annotation data. Both 2D and 3D auto-segmentation networks were evaluated for their performance. Furthermore, we evaluated the performance of semi-supervised method for 50 labeled data and only 10 labeled data.
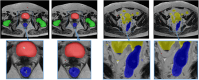
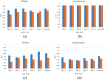
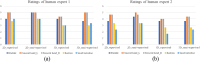
Results: The Dice similarity coefficient (DSC) of the bladder, femoral heads, rectum and small intestine between segmentation results and reference masks is 0.954, 0.984, 0.908, 0.852 only using self-annotation and post-processing methods of 2D segmentation model. The DSC of corresponding OARs is 0.871, 0.975, 0.975, 0.783, 0.724 using 3D segmentation network, 0.896, 0.984, 0.890, 0.828 using 2D segmentation network and common supervised method.
Conclusion: The outcomes of our study demonstrate that it is possible to train a multi-OAR segmentation model using small annotation samples and additional unlabeled data. To effectively annotate the dataset, ensemble learning and post-processing methods were employed. Additionally, when dealing with anisotropy and limited sample sizes, the 2D model outperformed the 3D model in terms of performance.
Keywords: auto-segmentation; deep-learning; semi-supervised learning.
© 2024 The Authors. Journal of Applied Clinical Medical Physics published by Wiley Periodicals, LLC on behalf of The American Association of Physicists in Medicine.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Semi-supervised abdominal multi-organ segmentation by object-redrawing.Med Phys. 2024 Nov;51(11):8334-8347. doi: 10.1002/mp.17364. Epub 2024 Aug 21. Med Phys. 2024. PMID: 39167059
-
Automatic delineation of cervical cancer target volumes in small samples based on multi-decoder and semi-supervised learning and clinical application.Sci Rep. 2024 Nov 6;14(1):26937. doi: 10.1038/s41598-024-78424-0. Sci Rep. 2024. PMID: 39505991 Free PMC article.
-
Semi-supervised learning framework with shape encoding for neonatal ventricular segmentation from 3D ultrasound.Med Phys. 2024 Sep;51(9):6134-6148. doi: 10.1002/mp.17242. Epub 2024 Jun 10. Med Phys. 2024. PMID: 38857570
-
Multi-organ segmentation: a progressive exploration of learning paradigms under scarce annotation.Phys Med Biol. 2024 May 14;69(11). doi: 10.1088/1361-6560/ad33b5. Phys Med Biol. 2024. PMID: 38479023 Review.
-
Auto-segmentation of organs at risk for head and neck radiotherapy planning: From atlas-based to deep learning methods.Med Phys. 2020 Sep;47(9):e929-e950. doi: 10.1002/mp.14320. Epub 2020 Jul 28. Med Phys. 2020. PMID: 32510603 Review.
Cited by
-
How much data do you need? An analysis of pelvic multi-organ segmentation in a limited data context.Phys Eng Sci Med. 2025 Mar;48(1):409-419. doi: 10.1007/s13246-024-01514-w. Epub 2025 Mar 11. Phys Eng Sci Med. 2025. PMID: 40067638 Free PMC article.
References
-
- Yang S‐D, Zhao Yu, Zhang F, et al. An efficient two‐step multi‐organ registration on abdominal CT via deep‐learning based segmentation. Biomed Signal Process Control. 2021;70:103027. doi:10.1016/j.bspc.2021.103027 - DOI
LinkOut - more resources
Full Text Sources

