Prolonged overexpression of PLK4 leads to formation of centriole rosette clusters that are connected via canonical centrosome linker proteins
- PMID: 38388511
- PMCID: PMC10883960
- DOI: 10.1038/s41598-024-53985-2
Prolonged overexpression of PLK4 leads to formation of centriole rosette clusters that are connected via canonical centrosome linker proteins
Abstract
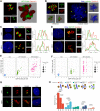
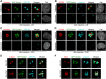
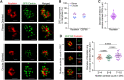
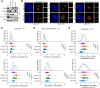
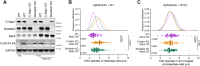
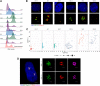
Centrosome amplification is a hallmark of cancer and PLK4 is one of the responsible factors for cancer associated centrosome amplification. Increased PLK4 levels was also shown to contribute to generation of cells with centriole amplification in mammalian tissues as olfactory neuron progenitor cells. PLK4 overexpression generates centriole rosette (CR) structures which harbor more than two centrioles each. Long term PLK4 overexpression results with centrosome amplification, but the maturation of amplified centrioles in CRs and linking of PLK4 induced amplified centrosomes has not yet been investigated in detail. Here, we show evidence for generation of large clustered centrosomes which have more than 2 centriole rosettes and define these structures as centriole rosette clusters (CRCs) in cells that have high PLK4 levels for 2 consecutive cell cycles. In addition, we show that PLK4 induced CRs follow normal centrosomal maturation processes and generate CRC structures that are inter-connected with canonical centrosomal linker proteins as C-Nap1, Rootletin and Cep68 in the second cell cycle after PLK4 induction. Increased PLK4 levels in cells with C-Nap1 and Rootletin knock-out resulted with distanced CRs and CRCs in interphase, while Nek2 knock-out inhibited separation of CRCs in prometaphase, providing functional evidence for the binding of CRC structures with centrosomal linker proteins. Taken together, these results suggest a cell cycle dependent model for PLK4 induced centrosome amplification which occurs in 2 consecutive cell cycles: (i) CR state in the first cell cycle, and (ii) CRC state in the second cell cycle.
Keywords: Centrosome amplification; Centrosome linkers; PLK4.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Centriole splitting caused by loss of the centrosomal linker protein C-NAP1 reduces centriolar satellite density and impedes centrosome amplification.Mol Biol Cell. 2017 Mar 15;28(6):736-745. doi: 10.1091/mbc.E16-05-0325. Epub 2017 Jan 18. Mol Biol Cell. 2017. PMID: 28100636 Free PMC article.
-
Cep78 is a new centriolar protein involved in Plk4-induced centriole overduplication.J Cell Sci. 2016 Jul 15;129(14):2713-8. doi: 10.1242/jcs.184093. Epub 2016 May 31. J Cell Sci. 2016. PMID: 27246242
-
The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4.Curr Biol. 2009 Jan 13;19(1):43-9. doi: 10.1016/j.cub.2008.11.037. Epub 2008 Dec 11. Curr Biol. 2009. PMID: 19084407
-
Role of Polo-like Kinases Plk1 and Plk4 in the Initiation of Centriole Duplication-Impact on Cancer.Cells. 2022 Feb 24;11(5):786. doi: 10.3390/cells11050786. Cells. 2022. PMID: 35269408 Free PMC article. Review.
-
The centrosome duplication cycle in health and disease.FEBS Lett. 2014 Aug 1;588(15):2366-72. doi: 10.1016/j.febslet.2014.06.030. Epub 2014 Jun 18. FEBS Lett. 2014. PMID: 24951839 Review.
Cited by
-
Nek2A prevents centrosome clustering and induces cell death in cancer cells via KIF2C interaction.Cell Death Dis. 2024 Mar 16;15(3):222. doi: 10.1038/s41419-024-06601-0. Cell Death Dis. 2024. PMID: 38493150 Free PMC article.
-
Influence of Hypoxia on the Airway Epithelium.Physiol Res. 2024 Nov 29;73(S2):S557. doi: 10.33549/physiolres.935436. Physiol Res. 2024. PMID: 39589303 Free PMC article. Review.
References
-
- Reiter JF, Leroux MR. Cilia in development: The coming of age of a vibrant research field. Development. 2017;144:733–737. - PubMed
-
- Nigg EA. The centrosome in development and disease. Chromosome Res. 2002;10:3–11.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

