M2 macrophages independently promote beige adipogenesis via blocking adipocyte Ets1
- PMID: 38388532
- PMCID: PMC10883921
- DOI: 10.1038/s41467-024-45899-4
M2 macrophages independently promote beige adipogenesis via blocking adipocyte Ets1
Abstract
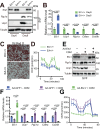
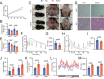
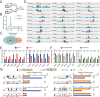
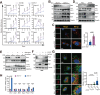
Adipose tissue macrophages can promote beige adipose thermogenesis by altering local sympathetic activity. Here, we perform sympathectomy in mice and further eradicate subcutaneous adipose macrophages and discover that these macrophages have a direct beige-promoting function that is independent of sympathetic system. We further identify adipocyte Ets1 as a vital mediator in this process. The anti-inflammatory M2 macrophages suppress Ets1 expression in adipocytes, transcriptionally activate mitochondrial biogenesis, as well as suppress mitochondrial clearance, thereby increasing the mitochondrial numbers and promoting the beiging process. Male adipocyte Ets1 knock-in mice are completely cold intolerant, whereas male mice lacking Ets1 in adipocytes show enhanced energy expenditure and are resistant to metabolic disorders caused by high-fat-diet. Our findings elucidate a direct communication between M2 macrophages and adipocytes, and uncover a function for Ets1 in responding to macrophages and negatively governing mitochondrial content and beige adipocyte formation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Foxp1 controls brown/beige adipocyte differentiation and thermogenesis through regulating β3-AR desensitization.Nat Commun. 2019 Nov 7;10(1):5070. doi: 10.1038/s41467-019-12988-8. Nat Commun. 2019. PMID: 31699980 Free PMC article.
-
miR-130b/301b Is a Negative Regulator of Beige Adipogenesis and Energy Metabolism In Vitro and In Vivo.Diabetes. 2022 Nov 1;71(11):2360-2371. doi: 10.2337/db22-0205. Diabetes. 2022. PMID: 36001751 Free PMC article.
-
Overexpression of Adiponectin Receptor 1 Inhibits Brown and Beige Adipose Tissue Activity in Mice.Int J Mol Sci. 2021 Jan 18;22(2):906. doi: 10.3390/ijms22020906. Int J Mol Sci. 2021. PMID: 33477525 Free PMC article.
-
Critical review of beige adipocyte thermogenic activation and contribution to whole-body energy expenditure.Horm Mol Biol Clin Investig. 2017 Sep 1;31(2):/j/hmbci.2017.31.issue-2/hmbci-2017-0042/hmbci-2017-0042.xml. doi: 10.1515/hmbci-2017-0042. Horm Mol Biol Clin Investig. 2017. PMID: 28862985 Review.
-
Intercellular and inter-organ crosstalk in browning of white adipose tissue: molecular mechanism and therapeutic complications.J Mol Cell Biol. 2021 Oct 21;13(7):466-479. doi: 10.1093/jmcb/mjab038. J Mol Cell Biol. 2021. PMID: 34185049 Free PMC article. Review.
Cited by
-
ETS emerges to heat up adipose.Genes Dev. 2025 Jul 1;39(13-14):789-791. doi: 10.1101/gad.352966.125. Genes Dev. 2025. PMID: 40389324 Free PMC article. Review.
-
Peripheral nerve-derived CSF1 induces BMP2 expression in macrophages to promote nerve regeneration and wound healing.NPJ Regen Med. 2024 Nov 21;9(1):35. doi: 10.1038/s41536-024-00379-7. NPJ Regen Med. 2024. PMID: 39572537 Free PMC article.
-
Transcriptional control of metabolism by interferon regulatory factors.Nat Rev Endocrinol. 2024 Oct;20(10):573-587. doi: 10.1038/s41574-024-00990-0. Epub 2024 May 20. Nat Rev Endocrinol. 2024. PMID: 38769435 Free PMC article. Review.
-
Electron-donable heterojunctions with synergetic Ru-Cu pair sites for biocatalytic microenvironment modulations in inflammatory mandible defects.Nat Commun. 2024 Nov 6;15(1):9592. doi: 10.1038/s41467-024-53824-y. Nat Commun. 2024. PMID: 39505847 Free PMC article.
-
Role of the E26 transformation specific transcription factor family in metabolic disorders.J Endocrinol Invest. 2025 Jul 5. doi: 10.1007/s40618-025-02634-0. Online ahead of print. J Endocrinol Invest. 2025. PMID: 40616738 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

