A novel ferroptosis-related gene signature for overall survival prediction in patients with gastric cancer
- PMID: 38388534
- PMCID: PMC10883968
- DOI: 10.1038/s41598-024-53515-0
A novel ferroptosis-related gene signature for overall survival prediction in patients with gastric cancer
Abstract
The global diagnosis rate and mortality of gastric cancer (GC) are among the highest. Ferroptosis and iron-metabolism have a profound impact on tumor development and are closely linked to cancer treatment and patient's prognosis. In this study, we identified six PRDEGs (prognostic ferroptosis- and iron metabolism-related differentially expressed genes) using LASSO-penalized Cox regression analysis. The TCGA cohort was used to establish a prognostic risk model, which allowed us to categorize GC patients into the high- and the low-risk groups based on the median value of the risk scores. Our study demonstrated that patients in the low-risk group had a higher probability of survival compared to those in the high-risk group. Furthermore, the low-risk group exhibited a higher tumor mutation burden (TMB) and a longer 5-year survival period when compared to the high-risk group. In summary, the prognostic risk model, based on the six genes associated with ferroptosis and iron-metabolism, performs well in predicting the prognosis of GC patients.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

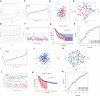
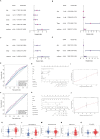



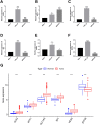
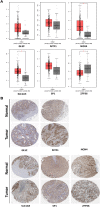
References
MeSH terms
Substances
Grants and funding
- No.82204641/National Outstanding Youth Science Fund Project of National Natural Science Foundation of China
- No. 2022-239/The Fifth Batch of National Traditional Chinese Medicine Excellent Clinical Talents Training Project (Announcement from the Personnel and Education Department of the National Administration of Traditional Chinese Medicine.)
- No.XPT82204641/Nanjing University of Chinese Medicine National Natural Science Foundation Young Scientists Fund Project Matching Fund Project
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

