Genome-wide detection of positive and balancing signatures of selection shared by four domesticated rainbow trout populations (Oncorhynchus mykiss)
- PMID: 38389056
- PMCID: PMC10882880
- DOI: 10.1186/s12711-024-00884-9
Genome-wide detection of positive and balancing signatures of selection shared by four domesticated rainbow trout populations (Oncorhynchus mykiss)
Abstract
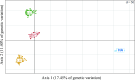
Background: Evolutionary processes leave footprints along the genome over time. Highly homozygous regions may correspond to positive selection of favorable alleles, while maintenance of heterozygous regions may be due to balancing selection phenomena. We analyzed data from 176 fish from four disconnected domestic rainbow trout populations that were genotyped using a high-density Axiom Trout genotyping 665K single nucleotide polymorphism array, including 20 from the US and 156 from three French lines. Using methods based on runs of homozygosity and extended haplotype homozygosity, we detected signatures of selection in these four populations.
Results: Nine genomic regions that included 253 genes were identified as being under positive selection in all four populations Most were located on chromosome 2 but also on chromosomes 12, 15, 16, and 20. In addition, four heterozygous regions that contain 29 genes that are putatively under balancing selection were also shared by the four populations. These were located on chromosomes 10, 13, and 19. Regardless of the homozygous or heterozygous nature of the regions, in each region, we detected several genes that are highly conserved among vertebrates due to their critical roles in cellular and nuclear organization, embryonic development, or immunity. We identified new candidate genes involved in rainbow trout fitness, as well as 17 genes that were previously identified to be under positive selection, 10 of which in other fishes (auts2, atp1b3, zp4, znf135, igf-1α, brd2, col9a2, mrap2, pbx1, and emilin-3).
Conclusions: Using material from disconnected populations of different origins allowed us to draw a genome-wide map of signatures of positive selection that are shared between these rainbow trout populations, and to identify several regions that are putatively under balancing selection. These results provide a valuable resource for future investigations of the dynamics of genetic diversity and genome evolution during domestication.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Lynch M, Conery J, Burger R. Mutation accumulation and the extinction of small populations. Am Nat. 1995;146:489–518.
-
- East EM. The role of reproduction in evolution. Am Nat. 1918;52:273–289.
-
- Fisher RA. The genetical theory of natural selection. Oxford: Oxford University Press; 1958.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

