Pig pangenome graph reveals functional features of non-reference sequences
- PMID: 38389084
- PMCID: PMC10882747
- DOI: 10.1186/s40104-023-00984-4
Pig pangenome graph reveals functional features of non-reference sequences
Abstract
Background: The reliance on a solitary linear reference genome has imposed a significant constraint on our comprehensive understanding of genetic variation in animals. This constraint is particularly pronounced for non-reference sequences (NRSs), which have not been extensively studied.
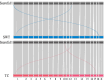
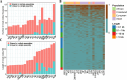
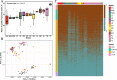
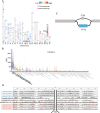
Results: In this study, we constructed a pig pangenome graph using 21 pig assemblies and identified 23,831 NRSs with a total length of 105 Mb. Our findings revealed that NRSs were more prevalent in breeds exhibiting greater genetic divergence from the reference genome. Furthermore, we observed that NRSs were rarely found within coding sequences, while NRS insertions were enriched in immune-related Gene Ontology terms. Notably, our investigation also unveiled a close association between novel genes and the immune capacity of pigs. We observed substantial differences in terms of frequencies of NRSs between Eastern and Western pigs, and the heat-resistant pigs exhibited a substantial number of NRS insertions in an 11.6 Mb interval on chromosome X. Additionally, we discovered a 665 bp insertion in the fourth intron of the TNFRSF19 gene that may be associated with the ability of heat tolerance in Southern Chinese pigs.
Conclusions: Our findings demonstrate the potential of a graph genome approach to reveal important functional features of NRSs in pig populations.
Keywords: Heat tolerance; Immune ability; Non-reference sequences; Pig pangenome.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Pangenome graph mitigates heterozygosity overestimation from mapping bias: a case study in Chinese indigenous pigs.BMC Biol. 2025 Mar 26;23(1):89. doi: 10.1186/s12915-025-02194-y. BMC Biol. 2025. PMID: 40140905 Free PMC article.
-
Human pangenome analysis of sequences missing from the reference genome reveals their widespread evolutionary, phenotypic, and functional roles.Nucleic Acids Res. 2024 Mar 21;52(5):2212-2230. doi: 10.1093/nar/gkae086. Nucleic Acids Res. 2024. PMID: 38364871 Free PMC article.
-
Pangenome analysis reveals transposon-driven genome evolution in cotton.BMC Biol. 2024 Apr 23;22(1):92. doi: 10.1186/s12915-024-01893-2. BMC Biol. 2024. PMID: 38654264 Free PMC article.
-
Chromosome-level Genome Assembly of Korean Long-tailed Chicken and Pangenome of 40 Gallus gallus Assemblies.Sci Data. 2025 Jan 11;12(1):51. doi: 10.1038/s41597-024-04287-9. Sci Data. 2025. PMID: 39799174 Free PMC article.
-
A review of the pangenome: how it affects our understanding of genomic variation, selection and breeding in domestic animals?J Anim Sci Biotechnol. 2023 May 5;14(1):73. doi: 10.1186/s40104-023-00860-1. J Anim Sci Biotechnol. 2023. PMID: 37143156 Free PMC article. Review.
Cited by
-
Pangenome graph mitigates heterozygosity overestimation from mapping bias: a case study in Chinese indigenous pigs.BMC Biol. 2025 Mar 26;23(1):89. doi: 10.1186/s12915-025-02194-y. BMC Biol. 2025. PMID: 40140905 Free PMC article.
-
Notable challenges posed by long-read sequencing for the study of transcriptional diversity and genome annotation.Genome Res. 2025 Apr 14;35(4):583-592. doi: 10.1101/gr.279865.124. Genome Res. 2025. PMID: 40032585 Free PMC article. Review.
-
Integrating parental genomes to reduce reference bias and identify intramuscular fat genes in Qinchuan Black pigs.J Anim Sci Biotechnol. 2025 Jul 20;16(1):104. doi: 10.1186/s40104-025-01236-3. J Anim Sci Biotechnol. 2025. PMID: 40684225 Free PMC article.
References
-
- Kehr B, Helgadottir A, Melsted P, Jonsson H, Helgason H, Jonasdottir A, et al. Diversity in non-repetitive human sequences not found in the reference genome. Nat Genet. 2017;49:588–593. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

