Microbiome structure variation and soybean's defense responses during flooding stress and elevated CO2
- PMID: 38389716
- PMCID: PMC10882081
- DOI: 10.3389/fpls.2023.1295674
Microbiome structure variation and soybean's defense responses during flooding stress and elevated CO2
Abstract
Introduction: With current trends in global climate change, both flooding episodes and higher levels of CO2 have been key factors to impact plant growth and stress tolerance. Very little is known about how both factors can influence the microbiome diversity and function, especially in tolerant soybean cultivars. This work aims to (i) elucidate the impact of flooding stress and increased levels of CO2 on the plant defenses and (ii) understand the microbiome diversity during flooding stress and elevated CO2 (eCO2).
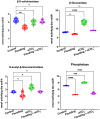
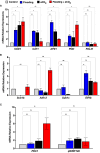
Methods: We used next-generation sequencing and bioinformatic methods to show the impact of natural flooding and eCO2 on the microbiome architecture of soybean plants' below- (soil) and above-ground organs (root and shoot). We used high throughput rhizospheric extra-cellular enzymes and molecular analysis of plant defense-related genes to understand microbial diversity in plant responses during eCO2 and flooding.
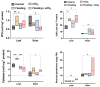
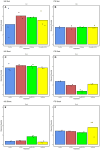
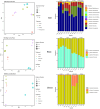
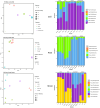
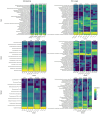
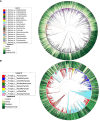
Results: Results revealed that bacterial and fungal diversity was substantially higher in combined flooding and eCO2 treatments than in non-flooding control. Microbial diversity was soil>root>shoot in response to flooding and eCO2. We found that sole treatment of eCO2 and flooding had significant abundances of Chitinophaga, Clostridium, and Bacillus. Whereas the combination of flooding and eCO2 conditions showed a significant abundance of Trichoderma and Gibberella. Rhizospheric extra-cellular enzyme activities were significantly higher in eCO2 than flooding or its combination with eCO2. Plant defense responses were significantly regulated by the oxidative stress enzyme activities and gene expression of Elongation factor 1 and Alcohol dehydrogenase 2 in floodings and eCO2 treatments in soybean plant root or shoot parts.
Conclusion: This work suggests that climatic-induced changes in eCO2 and submergence can reshape microbiome structure and host defenses, essential in plant breeding and developing stress-tolerant crops. This work can help in identifying core-microbiome species that are unique to flooding stress environments and increasing eCO2.
Keywords: climatic CO2; diversity; flooding stress; gene expression; microbiome; oxidative stress; soybean.
Copyright © 2024 Coffman, Mejia, Alicea, Mustafa, Ahmad, Crawford and Khan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Flooding episodes and seed treatment influence the microbiome diversity and function in the soybean root and rhizosphere.Sci Total Environ. 2025 Jun 20;982:179554. doi: 10.1016/j.scitotenv.2025.179554. Epub 2025 May 13. Sci Total Environ. 2025. PMID: 40367854
-
The silicon regulates microbiome diversity and plant defenses during cold stress in Glycine max L.Front Plant Sci. 2024 Jan 10;14:1280251. doi: 10.3389/fpls.2023.1280251. eCollection 2023. Front Plant Sci. 2024. PMID: 38269137 Free PMC article.
-
Tree growth rate regulate the influence of elevated CO2 on soil biochemical responses under tropical condition.J Environ Manage. 2019 Feb 1;231:1211-1221. doi: 10.1016/j.jenvman.2018.11.025. Epub 2018 Nov 17. J Environ Manage. 2019. PMID: 30602246
-
A meta-analysis of the combined effects of elevated carbon dioxide and chronic warming on plant %N, protein content and N-uptake rate.AoB Plants. 2021 May 25;13(4):plab031. doi: 10.1093/aobpla/plab031. eCollection 2021 Aug. AoB Plants. 2021. PMID: 34285792 Free PMC article. Review.
-
Salicylic acid and jasmonic acid in elevated CO2-induced plant defense response to pathogens.J Plant Physiol. 2023 Jul;286:154019. doi: 10.1016/j.jplph.2023.154019. Epub 2023 May 20. J Plant Physiol. 2023. PMID: 37244001 Review.
Cited by
-
CRISPR/Cas9: efficient and emerging scope for Brassica crop improvement.Planta. 2025 Jun 4;262(1):14. doi: 10.1007/s00425-025-04727-9. Planta. 2025. PMID: 40464976 Review.
-
Trichoderma: a multifunctional agent in plant health and microbiome interactions.BMC Microbiol. 2025 Jul 12;25(1):434. doi: 10.1186/s12866-025-04158-2. BMC Microbiol. 2025. PMID: 40652165 Free PMC article. Review.
References
-
- Aebi H. (1984). “[13] catalase in vitro ,” in Methods in enzymology (Amsterdam, Netherland: Elsevier; ), 121–126. - PubMed
-
- Ahmad P., Prasad M. N. V. (2011). Environmental adaptations and stress tolerance of plants in the era of climate change (New York, NY: Springer Science & Business Media; ).
LinkOut - more resources
Full Text Sources

