High-content stimulated Raman histology of human breast cancer
- PMID: 38389847
- PMCID: PMC10879861
- DOI: 10.7150/thno.90336
High-content stimulated Raman histology of human breast cancer
Abstract
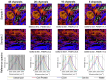
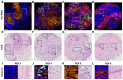
Histological examination is crucial for cancer diagnosis, however, the labor-intensive sample preparation involved in the histology impedes the speed of diagnosis. Recently developed two-color stimulated Raman histology could bypass the complex tissue processing to generates result close to hematoxylin and eosin staining, which is one of the golden standards in cancer histology. Yet, the underlying chemical features are not revealed in two-color stimulated Raman histology, compromising the effectiveness of prognostic stratification. Here, we present a high-content stimulated Raman histology (HC-SRH) platform that provides both morphological and chemical information for cancer diagnosis based on un-stained breast tissues. Methods: By utilizing both hyperspectral SRS imaging in the C-H vibration window and sparsity-penalized unmixing of overlapped spectral profiles, HC-SRH enabled high-content chemical mapping of saturated lipids, unsaturated lipids, cellular protein, extracellular matrix (ECM), and water. Spectral selective sampling was further implemented to boost the speed of HC-SRH. To show the potential for clinical use, HC-SRH using a compact fiber laser-based stimulated Raman microscope was demonstrated. Harnessing the wide and rapid tuning capability of the fiber laser, both C-H and fingerprint vibration windows were accessed. Results: HC-SRH successfully mapped unsaturated lipids, cellular protein, extracellular matrix, saturated lipid, and water in breast tissue. With these five chemical maps, HC-SRH provided distinct contrast for tissue components including duct, stroma, fat cell, necrosis, and vessel. With selective spectral sampling, the speed of HC-SRH was improved by one order of magnitude. The fiber-laser-based HC-SRH produced the same image quality in the C-H window as the state-of-the-art solid laser. In the fingerprint window, nucleic acid and solid-state ester contrast was demonstrated. Conclusions: HC-SRH provides both morphological and chemical information of tissue in a label-free manner. The chemical information detected is beyond the reach of traditional hematoxylin and eosin staining and heralds the potential of HC-SRH for biomarker discovery.
Keywords: breast cancer; fiber laser; high-content chemical imaging; label-free histology; stimulated Raman histology.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Giaquinto AN, Sung H, Miller KD, Kramer JL, Newman LA, Minihan A. et al. Breast Cancer Statistics, 2022. CA Cancer J Clin. 2022;72:524–41. - PubMed
-
- Lazaro-Pacheco D, Shaaban AM, Rehman S, Rehman I. Raman spectroscopy of breast cancer. Applied Spectroscopy Reviews. 2020;55:439–75.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

