Highly potent and selective phosphatidylinositol 4-kinase IIIβ inhibitors as broad-spectrum anti-rhinoviral agents
- PMID: 38389877
- PMCID: PMC10880896
- DOI: 10.1039/d3md00630a
Highly potent and selective phosphatidylinositol 4-kinase IIIβ inhibitors as broad-spectrum anti-rhinoviral agents
Erratum in
-
Correction: Highly potent and selective phosphatidylinositol 4-kinase IIIβ inhibitors as broad-spectrum anti-rhinoviral agents.RSC Med Chem. 2024 Jun 12;15(6):2196-2197. doi: 10.1039/d4md90022g. eCollection 2024 Jun 19. RSC Med Chem. 2024. PMID: 38911159 Free PMC article.
Abstract
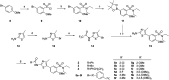
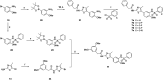
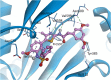
Human rhinoviruses (hRVs) cause upper and lower respiratory tract infections and exacerbate asthma and chronic obstructive pulmonary disease. hRVs comprise more than 160 strains with considerable genetic variation. Their high diversity and strain-specific interactions with antisera hinder the development of anti-hRV therapeutic agents. Phosphatidylinositol-4-kinase IIIβ (PI4KIIIβ) is a key enzyme in the phosphoinositide signalling pathway that is crucial for the replication and survival of various viruses. We identified novel PI4KIIIβ inhibitors, N-(4-methyl-5-arylthiazol)-2-amide derivatives, by generating a hit compound, 1a, from the high-throughput screening of a chemical library, followed by the optimization study of 1a. Inhibitor 7e exhibited the highest activity (EC50 = 0.008, 0.0068, and 0.0076 μM for hRV-B14, hRV-A16, and hRV-A21, respectively) and high toxicity (CC50 = 6.1 μM). Inhibitor 7f showed good activity and low toxicity and provided the highest selectivity index (SI ≥ 4638, >3116, and >2793 for hRV-B14, hRV-A16, and hRV-A21, respectively). Furthermore, 7f showed broad-spectrum activities against various hRVs, coxsackieviruses, and other enteroviruses, such as EV-A71 and EV-D68. The binding mode of the inhibitors was investigated using 7f, and the experimental results of plaque reduction, replicon and cytotoxicity, and time-of-drug-addition assays suggested that 7f acts as a PI4KIIIβ inhibitor. The kinase inhibition activity of this series of compounds against PI4KIIIα and PI4KIIIβ was assessed, and 7f demonstrated kinase inhibition activity with an IC50 value of 0.016 μM for PI4KIIIβ, but not for PI4KIIIα (>10 μM). Therefore, 7f represents a highly potent and selective PI4KIIIβ inhibitor for the further development of antiviral therapy against hRVs or other enteroviruses.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures

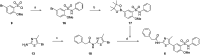
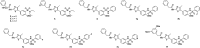



References
LinkOut - more resources
Full Text Sources
Chemical Information

