Inflammatory cytokines and oral lichen planus: a Mendelian randomization study
- PMID: 38390325
- PMCID: PMC10883046
- DOI: 10.3389/fimmu.2024.1332317
Inflammatory cytokines and oral lichen planus: a Mendelian randomization study
Abstract
Background: Inflammatory cytokines have long been considered closely related to the development of oral lichen planus (OLP), and we further explored the causal relationship between the two by Mendelian randomization (MR) method.
Methods: We performed bidirectional MR analyses by large genome-wide association studies (GWAS). The data included a large-scale OLP dataset, as well as datasets of 41 inflammatory cytokines. All data were obtained from the University of Bristol database, which includes 41 inflammatory cytokines, and the GWAS Catalog database, which includes 91 inflammatory cytokines. OLP data were obtained from the Finngen database, which includes 6411 cases and 405770 healthy controls. We used the inverse variance weighted (IVW) method, MR-Egger method, weighted median method, simple mode method and weighted mode method to analyze the causal relationship between inflammatory cytokines and OLP, and we also combined with sensitivity analysis to further verify the robustness of the results. We performed a meta-analysis of positive or potentially positive results for the same genes to confirm the reliability of the final results.
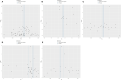
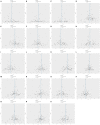
Results: We primarily used the IVW analysis method, corrected using the Benjamin Hochberg (BH) method. When p<0.00038 (0.05/132), the results are significantly causal; when 0.00038<p<0.05, the results are potentially causal. We found a total of 7 inflammatory cytokines with significant or potential associations with OLP (University of Bristol database: 2, GWAS Catalog database: 5). In the reverse analysis, we found that a total of 30 inflammatory cytokines were significantly or potentially associated with OLP (University of Bristol database: 5, GWAS Catalog database: 25). After sensitivity analysis and meta-analysis, we finally determined that there was a causal relationship between a total of 3 inflammatory cytokines and OLP in the forward analysis, the most significant of which was FGF21 (p=0.02954, odds ratio (OR): 1.113, 95% confidence interval (95%CI): 1.011-1.226). In the reverse analysis, 14 inflammatory cytokines were causally associated with OLP, the most significant of which was PLAU (p=0.00002, OR: 0.951, 95%CI: 0.930-0.973).
Conclusion: There is a causal association between OLP and some inflammatory cytokines, which may play an important role in the pathogenesis of OLP and require further attention.
Keywords: Mendelian randomization; immunity; inflammation; inflammatory cytokines; oral lichen planus.
Copyright © 2024 Chen, Zhang, Wu, Lei, Lei and Zhao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

