Examination of eQTL Polymorphisms Associated with Increased Risk of Progressive Complicated Sarcoidosis in European and African Descent Subjects
- PMID: 38390497
- PMCID: PMC10883688
Examination of eQTL Polymorphisms Associated with Increased Risk of Progressive Complicated Sarcoidosis in European and African Descent Subjects
Abstract
Background: A limited pool of SNPs are linked to the development and severity of sarcoidosis, a systemic granulomatous inflammatory disease. By integrating genome-wide association studies (GWAS) data and expression quantitative trait loci (eQTL) single nuclear polymorphisms (SNPs), we aimed to identify novel sarcoidosis SNPs potentially influencing the development of complicated sarcoidosis.
Methods: A GWAS (Affymetrix 6.0) involving 209 African-American (AA) and 193 European-American (EA, 75 and 51 complicated cases respectively) and publicly-available GWAS controls (GAIN) was utilized. Annotation of multi-tissue eQTL SNPs present on the GWAS created a pool of ~46,000 eQTL SNPs examined for association with sarcoidosis risk and severity (Logistic Model, Plink). The most significant EA/AA eQTL SNPs were genotyped in a sarcoidosis validation cohort (n=1034) and cross-validated in two independent GWAS cohorts.
Results: No single GWAS SNP achieved significance (p<1x10-8), however, analysis of the eQTL/GWAS SNP pool yielded 621 eQTL SNPs (p<10-4) associated with 730 genes that highlighted innate immunity, MHC Class II, and allograft rejection pathways with multiple SNPs validated in an independent sarcoidosis cohort (105 SNPs analyzed) (NOTCH4, IL27RA, BTNL2, ANXA11, HLA-DRB1). These studies confirm significant association of eQTL/GWAS SNPs in EAs and AAs with sarcoidosis risk and severity (complicated sarcoidosis) involving HLA region and innate immunity.
Conclusion: Despite the challenge of deciphering the genetic basis for sarcoidosis risk/severity, these results suggest that integrated eQTL/GWAS approaches may identify novel variants/genes and support the contribution of dysregulated innate immune responses to sarcoidosis severity.
Keywords: GWAS; SNPs; Sarcoidosis; expression quantitative trait loci (eQTL); polymorphism.
Conflict of interest statement
Competing interests Joe GN Garcia MD is CEO and Founder of Aqualung Therapeutics Corporation. David R. Moller MD is Chairman and CTO of Sarcoidosis Diagnostic Testing, LLC. All other authors report no conflict of interest.
Figures
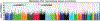

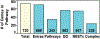


References
-
- Lockstone HE, Sanderson S, Kulakova N, Baban D, Leonard A, Kok WL, et al. Gene set analysis of lung samples provides insight into pathogenesis of progressive, fibrotic pulmonary sarcoidosis. Am J Respir Crit Care Med 2010; 181: 1367–1375. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
