Optimizing representations for integrative structural modeling using Bayesian model selection
- PMID: 38391029
- PMCID: PMC10924281
- DOI: 10.1093/bioinformatics/btae106
Optimizing representations for integrative structural modeling using Bayesian model selection
Abstract
Motivation: Integrative structural modeling combines data from experiments, physical principles, statistics of previous structures, and prior models to obtain structures of macromolecular assemblies that are challenging to characterize experimentally. The choice of model representation is a key decision in integrative modeling, as it dictates the accuracy of scoring, efficiency of sampling, and resolution of analysis. But currently, the choice is usually made ad hoc, manually.
Results: Here, we report NestOR (Nested Sampling for Optimizing Representation), a fully automated, statistically rigorous method based on Bayesian model selection to identify the optimal coarse-grained representation for a given integrative modeling setup. Given an integrative modeling setup, it determines the optimal representations from given candidate representations based on their model evidence and sampling efficiency. The performance of NestOR was evaluated on a benchmark of four macromolecular assemblies.
Availability and implementation: NestOR is implemented in the Integrative Modeling Platform (https://integrativemodeling.org) and is available at https://github.com/isblab/nestor. Data for the benchmark is at https://www.doi.org/10.5281/zenodo.10360718.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures
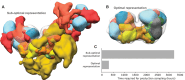
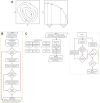

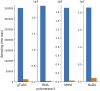
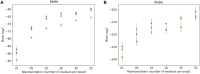
Update of
-
Optimizing representations for integrative structural modeling using Bayesian model selection.bioRxiv [Preprint]. 2023 Dec 13:2023.12.12.571227. doi: 10.1101/2023.12.12.571227. bioRxiv. 2023. Update in: Bioinformatics. 2024 Mar 4;40(3):btae106. doi: 10.1093/bioinformatics/btae106. PMID: 38168172 Free PMC article. Updated. Preprint.
References
-
- Alber F, Dokudovskaya S, Veenhoff LM. et al. Determining the architectures of macromolecular assemblies. Nature 2007;450:683–94. - PubMed
-
- Armache K-J, Mitterweger S, Meinhart A. et al. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J Biol Chem 2005;280:7131–4. - PubMed
-
- Ashton G, Bernstein N, Buchner J. et al. Nested sampling for physical scientists. Nat Rev Methods Primers 2022;2:39.

