Novel Biomarkers for Limb Girdle Muscular Dystrophy (LGMD)
- PMID: 38391941
- PMCID: PMC10886967
- DOI: 10.3390/cells13040329
Novel Biomarkers for Limb Girdle Muscular Dystrophy (LGMD)
Abstract
Objective: To identify novel biomarkers as an alternative diagnostic tool for limb girdle muscular dystrophy (LGMD).
Background: LGMD encompasses a group of muscular dystrophies characterized by proximal muscles weakness, elevated CK levels and dystrophic findings on muscle biopsy. Heterozygous CAPN3 mutations are associated with autosomal dominant LGMD-4, while biallelic mutations can cause autosomal recessive LGMD-1. Diagnosis is currently often based on invasive methods requiring muscle biopsy or blood tests. In most cases Western blotting (WB) analysis from muscle biopsy is essential for a diagnosis, as muscle samples are currently the only known tissues to express the full-length CAPN3 isoform.
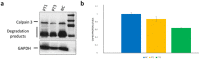
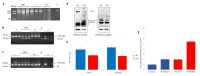
Methods: We analyzed CAPN3 in a cohort including 60 LGMD patients. Selected patients underwent a complete neurological examination, electromyography, muscle biopsy, and skin biopsies for primary fibroblasts isolation. The amount of CAPN3 was evaluated by WB analysis in muscle and skin tissues. The total RNA isolated from muscle, fibroblast and urine was processed, and cDNA was used for qualitative analysis. The expression of CAPN3 was investigated by qRT-PCR. The CAPN3 3D structure has been visualized and analyzed using PyMOL.
Results: Among our patients, seven different CAPN3 mutations were detected, of which two were novel. After sequencing CAPN3 transcripts from fibroblast and urine, we detected different CAPN3 isoforms surprisingly including the full-length transcript. We found comparable protein levels from fibroblasts and muscle tissue; in particular, patients harboring a novel CAPN3 mutation showed a 30% reduction in protein compared to controls from both tissues.
Conclusions: Our findings showed for the first time the presence of the CAPN3 full-length transcript in urine and skin samples. Moreover, we demonstrated surprisingly comparable CAPN3 protein levels between muscle and skin samples, thus allowing us to hypothesize the use of skin biopsy and probably of urine samples as an alternative less invasive method to assess the amount of CAPN3 when molecular diagnosis turns out to be inconclusive.
Keywords: CAPN3; RNA; limb girdle muscular dystrophy; western blotting analysis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Richard I., Broux O., Allamand V., Fougerousse F., Chiannilkulchai N., Bourg N., Brenguier L., Devaud C., Pasturaud P., Roudaut C., et al. Mutations in the Proteolytic Enzyme Calpain 3 Cause Limb-Girdle Muscular Dystrophy Type 2A. Cell. 1995;81:27–40. doi: 10.1016/0092-8674(95)90368-2. - DOI - PubMed
-
- Ermolova N., Kudryashova E., DiFranco M., Vergara J., Kramerova I., Spencer M.J. Pathogenity of Some Limb Girdle Muscular Dystrophy Mutations Can Result from Reduced Anchorage to Myofibrils and Altered Stability of Calpain 3. Hum. Mol. Genet. 2011;20:3331–3345. doi: 10.1093/hmg/ddr239. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

