Lateral Flow Biosensor for On-Site Multiplex Detection of Viruses Based on One-Step Reverse Transcription and Strand Displacement Amplification
- PMID: 38392022
- PMCID: PMC10886883
- DOI: 10.3390/bios14020103
Lateral Flow Biosensor for On-Site Multiplex Detection of Viruses Based on One-Step Reverse Transcription and Strand Displacement Amplification
Abstract
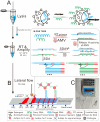
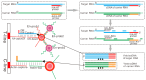
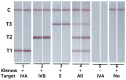
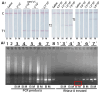
Respiratory pathogens pose a huge threat to public health, especially the highly mutant RNA viruses. Therefore, reliable, on-site, rapid diagnosis of such pathogens is an urgent need. Traditional assays such as nucleic acid amplification tests (NAATs) have good sensitivity and specificity, but these assays require complex sample pre-treatment and a long test time. Herein, we present an on-site biosensor for rapid and multiplex detection of RNA pathogens. Samples with viruses are first lysed in a lysis buffer containing carrier RNA to release the target RNAs. Then, the lysate is used for amplification by one-step reverse transcription and single-direction isothermal strand displacement amplification (SDA). The yield single-strand DNAs (ssDNAs) are visually detected by a lateral flow biosensor. With a secondary signal amplification system, as low as 20 copies/μL of virus can be detected in this study. This assay avoids the process of nucleic acid purification, making it equipment-independent and easier to operate, so it is more suitable for on-site molecular diagnostic applications.
Keywords: AuNPs; NAATs; RT-SDA; lateral flow biosensor; on-site detection.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Similar articles
-
Rapid and Visual Detection of SARS-CoV-2 Using Multiplex Reverse Transcription Loop-Mediated Isothermal Amplification Linked With Gold Nanoparticle-Based Lateral Flow Biosensor.Front Cell Infect Microbiol. 2021 Jul 14;11:581239. doi: 10.3389/fcimb.2021.581239. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 34336708 Free PMC article.
-
Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19.Biosens Bioelectron. 2020 Oct 15;166:112437. doi: 10.1016/j.bios.2020.112437. Epub 2020 Jul 15. Biosens Bioelectron. 2020. PMID: 32692666 Free PMC article.
-
Development of advanced control material for reverse transcription-mediated bacterial nucleic acid amplification tests.J Clin Microbiol. 2024 May 8;62(5):e0024324. doi: 10.1128/jcm.00243-24. Epub 2024 Apr 17. J Clin Microbiol. 2024. PMID: 38629844 Free PMC article.
-
Lateral flow biosensor for DNA extraction-free detection of Salmonella based on aptamer mediated strand displacement amplification.Biosens Bioelectron. 2014 Jun 15;56:192-7. doi: 10.1016/j.bios.2014.01.015. Epub 2014 Jan 17. Biosens Bioelectron. 2014. PMID: 24491961
-
Current Trends in RNA Virus Detection via Nucleic Acid Isothermal Amplification-Based Platforms.Biosensors (Basel). 2024 Feb 11;14(2):97. doi: 10.3390/bios14020097. Biosensors (Basel). 2024. PMID: 38392016 Free PMC article. Review.
Cited by
-
Simultaneously ultrasensitive and differential detection of SARS-CoV-2, adenovirus and influenza a virus using multiplex fluorescence lateral flow immunoassay.Front Immunol. 2025 May 9;16:1540676. doi: 10.3389/fimmu.2025.1540676. eCollection 2025. Front Immunol. 2025. PMID: 40416977 Free PMC article.
References
-
- Bhadra S., Jiang Y.S., Kumar M.R., Johnson R.F., Hensley L.E., Ellington A.D. Real-Time Sequence-Validated Loop-Mediated Isothermal Amplification Assays for Detection of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) PLoS ONE. 2015;10:e0123126. doi: 10.1371/journal.pone.0123126. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

