Role of Optimization in RNA-Protein-Binding Prediction
- PMID: 38392205
- PMCID: PMC11154364
- DOI: 10.3390/cimb46020087
Role of Optimization in RNA-Protein-Binding Prediction
Abstract
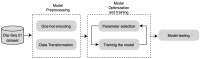
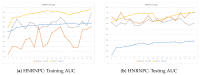
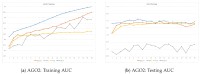
RNA-binding proteins (RBPs) play an important role in regulating biological processes, such as gene regulation. Understanding their behaviors, for example, their binding site, can be helpful in understanding RBP-related diseases. Studies have focused on predicting RNA binding by means of machine learning algorithms including deep convolutional neural network models. One of the integral parts of modeling deep learning is achieving optimal hyperparameter tuning and minimizing a loss function using optimization algorithms. In this paper, we investigate the role of optimization in the RBP classification problem using the CLIP-Seq 21 dataset. Three optimization methods are employed on the RNA-protein binding CNN prediction model; namely, grid search, random search, and Bayesian optimizer. The empirical results show an AUC of 94.42%, 93.78%, 93.23% and 92.68% on the ELAVL1C, ELAVL1B, ELAVL1A, and HNRNPC datasets, respectively, and a mean AUC of 85.30 on 24 datasets. This paper's findings provide evidence on the role of optimizers in improving the performance of RNA-protein binding prediction.
Keywords: Bayesian optimizer; RNA-binding proteins; artificial intelligence; bioinformatics; convolutional neural network (CNN); deep learning; grid search; machine learning; optimization; proteins; random search optimizer.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Improving classification accuracy of fine-tuned CNN models: Impact of hyperparameter optimization.Heliyon. 2024 Feb 23;10(5):e26586. doi: 10.1016/j.heliyon.2024.e26586. eCollection 2024 Mar 15. Heliyon. 2024. PMID: 38463880 Free PMC article.
-
Prediction of binding property of RNA-binding proteins using multi-sized filters and multi-modal deep convolutional neural network.PLoS One. 2019 Apr 26;14(4):e0216257. doi: 10.1371/journal.pone.0216257. eCollection 2019. PLoS One. 2019. PMID: 31026297 Free PMC article.
-
DeepPN: a deep parallel neural network based on convolutional neural network and graph convolutional network for predicting RNA-protein binding sites.BMC Bioinformatics. 2022 Jun 29;23(1):257. doi: 10.1186/s12859-022-04798-5. BMC Bioinformatics. 2022. PMID: 35768792 Free PMC article.
-
Artificial intelligence in clinical care amidst COVID-19 pandemic: A systematic review.Comput Struct Biotechnol J. 2021;19:2833-2850. doi: 10.1016/j.csbj.2021.05.010. Epub 2021 May 7. Comput Struct Biotechnol J. 2021. PMID: 34025952 Free PMC article. Review.
-
A survey on protein-DNA-binding sites in computational biology.Brief Funct Genomics. 2022 Sep 16;21(5):357-375. doi: 10.1093/bfgp/elac009. Brief Funct Genomics. 2022. PMID: 35652477 Review.
References
LinkOut - more resources
Full Text Sources

