General Overview of Klebsiella pneumonia: Epidemiology and the Role of Siderophores in Its Pathogenicity
- PMID: 38392297
- PMCID: PMC10886558
- DOI: 10.3390/biology13020078
General Overview of Klebsiella pneumonia: Epidemiology and the Role of Siderophores in Its Pathogenicity
Abstract
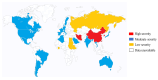
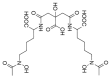
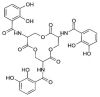
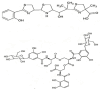
The opportunistic pathogen Klebsiella pneumoniae (K. pneumoniae) can colonize mucosal surfaces and spread from mucosae to other tissues, causing fatal infections. Medical equipment and the healthcare setting can become colonized by Klebsiella species, which are widely distributed in nature and can be found in water, soil, and animals. Moreover, a substantial number of community-acquired illnesses are also caused by this organism worldwide. These infections are characterized by a high rate of morbidity and mortality as well as the capacity to spread metastatically. Hypervirulent Klebsiella strains are thought to be connected to these infections. Four components are critical to this bacterium's pathogenicity-the capsule, lipopolysaccharide, fimbriae, and siderophores. Siderophores are secondary metabolites that allow iron to sequester from the surrounding medium and transport it to the intracellular compartment of the bacteria. A number of variables may lead to K. pneumoniae colonization in a specific area. Risk factors for infection include local healthcare practices, antibiotic use and misuse, infection control procedures, nutrition, gender, and age.
Keywords: Klebsiella pneumoniae; epidemiology; pathogenesis; siderophores.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- WHO . Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. World Health Organization; Geneva, Switzerland: 2017.
-
- Pomakova D.K., Hsiao C.-B., Beanan J.M., Olson R., MacDonald U., Keynan Y., Russo T.A. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: An emerging and under-recognized pathogenic variant. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:981–989. doi: 10.1007/s10096-011-1396-6. - DOI - PubMed
-
- Lan P., Shi Q., Zhang P., Chen Y., Yan R., Hua X., Jiang Y., Zhou J., Yu Y. Core genome allelic profiles of clinical Klebsiella pneumoniae strains using a random forest algorithm based on multilocus sequence typing scheme for hypervirulence analysis. J. Infect. Dis. 2020;221:263–271. doi: 10.1093/infdis/jiz562. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

