An Efficient Homologous Recombination-Based In Situ Protein-Labeling Method in Verticillium dahliae
- PMID: 38392300
- PMCID: PMC10886240
- DOI: 10.3390/biology13020081
An Efficient Homologous Recombination-Based In Situ Protein-Labeling Method in Verticillium dahliae
Abstract
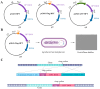
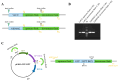
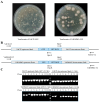
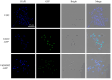
Accurate determination of protein localization, levels, or protein-protein interactions is pivotal for the study of their function, and in situ protein labeling via homologous recombination has emerged as a critical tool in many organisms. While this approach has been refined in various model fungi, the study of protein function in most plant pathogens has predominantly relied on ex situ or overexpression manipulations. To dissect the molecular mechanisms of development and infection for Verticillium dahliae, a formidable plant pathogen responsible for vascular wilt diseases, we have established a robust, homologous recombination-based in situ protein labeling strategy in this organism. Utilizing Agrobacterium tumefaciens-mediated transformation (ATMT), this methodology facilitates the precise tagging of specific proteins at their C-termini with epitopes, such as GFP and Flag, within the native context of V. dahliae. We demonstrate the efficacy of our approach through the in situ labeling of VdCf2 and VdDMM2, followed by subsequent confirmation via subcellular localization and protein-level analyses. Our findings confirm the applicability of homologous recombination for in situ protein labeling in V. dahliae and suggest its potential utility across a broad spectrum of filamentous fungi. This labeling method stands to significantly advance the field of functional genomics in plant pathogenic fungi, offering a versatile and powerful tool for the elucidation of protein function.
Keywords: Agrobacterium tumefaciens-mediated transformation (ATMT); Verticillium dahliae; filamentous fungi; homologous recombination; protein labeling.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures

Similar articles
-
An Improved Single-Step Cloning Strategy Simplifies the Agrobacterium tumefaciens-Mediated Transformation (ATMT)-Based Gene-Disruption Method for Verticillium dahliae.Phytopathology. 2016 Jun;106(6):645-52. doi: 10.1094/PHYTO-10-15-0280-R. Epub 2016 Mar 30. Phytopathology. 2016. PMID: 26780432
-
Functional analysis of autophagy genes via Agrobacterium-mediated transformation in the vascular Wilt fungus Verticillium dahliae.J Genet Genomics. 2013 Aug 20;40(8):421-31. doi: 10.1016/j.jgg.2013.04.006. Epub 2013 May 9. J Genet Genomics. 2013. PMID: 23969251
-
Protoplast transformation as a potential platform for exploring gene function in Verticillium dahliae.BMC Biotechnol. 2016 Jul 26;16(1):57. doi: 10.1186/s12896-016-0287-4. BMC Biotechnol. 2016. PMID: 27455996 Free PMC article.
-
An Overview of the Molecular Genetics of Plant Resistance to the Verticillium Wilt Pathogen Verticillium dahliae.Int J Mol Sci. 2020 Feb 7;21(3):1120. doi: 10.3390/ijms21031120. Int J Mol Sci. 2020. PMID: 32046212 Free PMC article. Review.
-
Interactions between Verticillium dahliae and cotton: pathogenic mechanism and cotton resistance mechanism to Verticillium wilt.Front Plant Sci. 2023 Apr 21;14:1174281. doi: 10.3389/fpls.2023.1174281. eCollection 2023. Front Plant Sci. 2023. PMID: 37152175 Free PMC article. Review.
References
-
- Snelders N.C., Rovenich H., Petti G.C., Rocafort M., van den Berg G.C.M., Vorholt J.A., Mesters J.R., Seidl M.F., Nijland R., Thomma B.P.H.J. Microbiome manipulation by a soil-borne fungal plant pathogen using effector proteins. Nat. Plants. 2020;6:1365–1374. doi: 10.1038/s41477-020-00799-5. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

