CXCL12+ dermal fibroblasts promote neutrophil recruitment and host defense by recognition of IL-17
- PMID: 38393304
- PMCID: PMC10890925
- DOI: 10.1084/jem.20231425
CXCL12+ dermal fibroblasts promote neutrophil recruitment and host defense by recognition of IL-17
Abstract
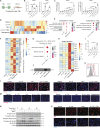
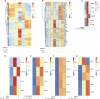
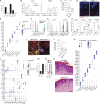
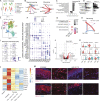
The skin provides an essential barrier for host defense through rapid action of multiple resident and recruited cell types, but the complex communication network governing these processes is incompletely understood. To define these cell-cell interactions more clearly, we performed an unbiased network analysis of mouse skin during invasive S. aureus infection and revealed a dominant role for CXCL12+ fibroblast subsets in neutrophil communication. These subsets predominantly reside in the reticular dermis, express adipocyte lineage markers, detect IL-17 and TNFα, and promote robust neutrophil recruitment through NFKBIZ-dependent release of CXCR2 ligands and CXCL12. Targeted deletion of Il17ra in mouse fibroblasts resulted in greatly reduced neutrophil recruitment and increased infection by S. aureus. Analogous human CXCL12+ fibroblast subsets abundantly express neutrophil chemotactic factors in psoriatic skin that are subsequently decreased upon therapeutic targeting of IL-17. These findings show that CXCL12+ dermal immune acting fibroblast subsets play a critical role in cutaneous neutrophil recruitment and host defense.
© 2024 Cavagnero et al.
Conflict of interest statement
Disclosures: R.L. Gallo is a cofounder and consultant of MatriSys Bioscience and has equity interest in this company. No other disclosures were reported.
Figures







References
-
- Balabanian, K., Lagane B., Infantino S., Chow K.Y., Harriague J., Moepps B., Arenzana-Seisdedos F., Thelen M., and Bachelerie F.. 2005. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J. Biol. Chem. 280:35760–35766. 10.1074/jbc.M508234200 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

