Serine/arginine-rich splicing factor 7 promotes the type I interferon response by activating Irf7 transcription
- PMID: 38393946
- PMCID: PMC11056844
- DOI: 10.1016/j.celrep.2024.113816
Serine/arginine-rich splicing factor 7 promotes the type I interferon response by activating Irf7 transcription
Abstract
Tight regulation of macrophage immune gene expression is required to fight infection without risking harmful inflammation. The contribution of RNA-binding proteins (RBPs) to shaping the macrophage response to pathogens remains poorly understood. Transcriptomic analysis reveals that a member of the serine/arginine-rich (SR) family of mRNA processing factors, SRSF7, is required for optimal expression of a cohort of interferon-stimulated genes in macrophages. Using genetic and biochemical assays, we discover that in addition to its canonical role in regulating alternative splicing, SRSF7 drives transcription of interferon regulatory transcription factor 7 (IRF7) to promote antiviral immunity. At the Irf7 promoter, SRSF7 maximizes STAT1 transcription factor binding and RNA polymerase II elongation via cooperation with the H4K20me1 histone methyltransferase KMT5a (SET8). These studies define a role for an SR protein in activating transcription and reveal an RBP-chromatin network that orchestrates macrophage antiviral gene expression.
Keywords: CP: Cell biology; CP: Molecular biology; RNA binding proteins; antiviral immunity; chromatin; co-transcriptional splicing; histone methylation; innate immunity; pre-mRNA splicing.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



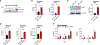


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

