Untranslated yet indispensable-UTRs act as key regulators in the environmental control of gene expression
- PMID: 38394144
- PMCID: PMC11263492
- DOI: 10.1093/jxb/erae073
Untranslated yet indispensable-UTRs act as key regulators in the environmental control of gene expression
Abstract
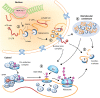
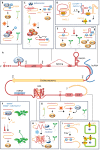
To survive and thrive in a dynamic environment, plants must continuously monitor their surroundings and adjust their development and physiology accordingly. Changes in gene expression underlie these developmental and physiological adjustments, and are traditionally attributed to widespread transcriptional reprogramming. Growing evidence, however, suggests that post-transcriptional mechanisms also play a vital role in tailoring gene expression to a plant's environment. Untranslated regions (UTRs) act as regulatory hubs for post-transcriptional control, harbouring cis-elements that affect an mRNA's processing, localization, translation, and stability, and thereby tune the abundance of the encoded protein. Here, we review recent advances made in understanding the critical function UTRs exert in the post-transcriptional control of gene expression in the context of a plant's abiotic environment. We summarize the molecular mechanisms at play, present examples of UTR-controlled signalling cascades, and discuss the potential that resides within UTRs to render plants more resilient to a changing climate.
Keywords: Abiotic stress; RNA processing; RNA structure; RNA-binding protein (RBP); alternative splicing; gene expression; post-transcriptional regulation; translation; untranslated region (UTR).
© The Author(s) 2024. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Anderson SJ, Kramer MC, Gosai SJ, et al.. 2018. N6-methyladenosine inhibits local ribonucleolytic cleavage to stabilize mRNAs in Arabidopsis. Cell Reports 25, 1146–1157.e3. - PubMed
-
- Artz O, Ackermann A, Taylor L, Koo PK, Pedmale UV.. 2023. Light and temperature regulate m6A-RNA modification to regulate growth in plants. bioRxiv doi: 10.1101/2023.01.17.524395. [Preprint]. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

