Roles of RNA scaffolding in nanoscale Gag multimerization and selective protein sorting at HIV membranes
- PMID: 38394201
- PMCID: PMC10889351
- DOI: 10.1126/sciadv.adk8297
Roles of RNA scaffolding in nanoscale Gag multimerization and selective protein sorting at HIV membranes
Abstract
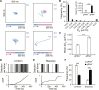
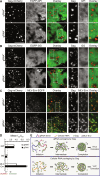
HIV-1 Gag proteins can multimerize upon the viral genomic RNA or multiple random cellular messenger RNAs to form a virus particle or a virus-like particle, respectively. To date, whether the two types of particles form via the same Gag multimerization process has remained unclarified. Using photoactivated localization microscopy to illuminate Gag organizations and dynamics at the nanoscale, here, we showed that genomic RNA mediates Gag multimerization in a more cluster-centric, cooperative, and spatiotemporally coordinated fashion, with the ability to drive dense Gag clustering dependent on its ability to act as a long-stranded scaffold not easily attainable by cellular messenger RNAs. These differences in Gag multimerization were further shown to affect downstream selective protein sorting into HIV membranes, indicating that the choice of RNA for packaging can modulate viral membrane compositions. These findings should advance the understanding of HIV assembly and further benefit the development of virus-like particle-based therapeutics.
Figures


References
-
- Bieniasz P., Telesnitsky A., Multiple, switchable protein:RNA interactions regulate human immunodeficiency virus type 1 assembly. Annu. Rev. Virol. 5, 165–183 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

