Therapeutic targeting Tudor domains in leukemia via CRISPR-Scan Assisted Drug Discovery
- PMID: 38394203
- PMCID: PMC10889360
- DOI: 10.1126/sciadv.adk3127
Therapeutic targeting Tudor domains in leukemia via CRISPR-Scan Assisted Drug Discovery
Abstract
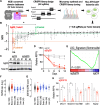
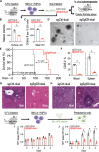
Epigenetic dysregulation has been reported in multiple cancers including leukemias. Nonetheless, the roles of the epigenetic reader Tudor domains in leukemia progression and therapy remain unexplored. Here, we conducted a Tudor domain-focused CRISPR screen and identified SGF29, a component of SAGA/ATAC acetyltransferase complexes, as a crucial factor for H3K9 acetylation, ribosomal gene expression, and leukemogenesis. To facilitate drug development, we integrated the CRISPR tiling scan with compound docking and molecular dynamics simulation, presenting a generally applicable strategy called CRISPR-Scan Assisted Drug Discovery (CRISPR-SADD). Using this approach, we identified a lead inhibitor that selectively targets SGF29's Tudor domain and demonstrates efficacy against leukemia. Furthermore, we propose that the structural genetics approach used in our study can be widely applied to diverse fields for de novo drug discovery.
Figures





References
-
- Dohner H., Weisdorf D. J., Bloomfield C. D., Acute myeloid leukemia. N. Engl. J. Med. 373, 1136–1152 (2015). - PubMed
-
- Shallis R. M., Wang R., Davidoff A., Ma X., Zeidan A. M., Epidemiology of acute myeloid leukemia: Recent progress and enduring challenges. Blood Rev. 36, 70–87 (2019). - PubMed
-
- Gu Z., Churchman M. L., Roberts K. G., Moore I., Zhou X., Nakitandwe J., Hagiwara K., Pelletier S., Gingras S., Berns H., Payne-Turner D., Hill A., Iacobucci I., Shi L., Pounds S., Cheng C., Pei D., Qu C., Newman S., Devidas M., Dai Y., Reshmi S. C., Gastier-Foster J., Raetz E. A., Borowitz M. J., Wood B. L., Carroll W. L., Zweidler-McKay P. A., Rabin K. R., Mattano L. A., Maloney K. W., Rambaldi A., Spinelli O., Radich J. P., Minden M. D., Rowe J. M., Luger S., Litzow M. R., Tallman M. S., Racevskis J., Zhang Y., Bhatia R., Kohlschmidt J., Mrozek K., Bloomfield C. D., Stock W., Kornblau S., Kantarjian H. M., Konopleva M., Evans W. E., Jeha S., Pui C. H., Yang J., Paietta E., Downing J. R., Relling M. V., Zhang J., Loh M. L., Hunger S. P., Mullighan C. G., PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat. Genet. 51, 296–307 (2019). - PMC - PubMed
-
- Chen C. W., Koche R. P., Sinha A. U., Deshpande A. J., Zhu N., Eng R., Doench J. G., Xu H., Chu S. H., Qi J., Wang X., Delaney C., Bernt K. M., Root D. E., Hahn W. C., Bradner J. E., Armstrong S. A., DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia. Nat. Med. 21, 335–343 (2015). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA236399/CA/NCI NIH HHS/United States
- P50 CA206963/CA/NCI NIH HHS/United States
- R01 CA259273/CA/NCI NIH HHS/United States
- R01 CA278050/CA/NCI NIH HHS/United States
- R01 CA214965/CA/NCI NIH HHS/United States
- P01 CA066996/CA/NCI NIH HHS/United States
- R37 CA233691/CA/NCI NIH HHS/United States
- P30 CA033572/CA/NCI NIH HHS/United States
- R01 CA243386/CA/NCI NIH HHS/United States
- R01 CA236626/CA/NCI NIH HHS/United States
- U54 CA243124/CA/NCI NIH HHS/United States
- R01 CA271497/CA/NCI NIH HHS/United States
- R01 CA280389/CA/NCI NIH HHS/United States
- R01 GM117923/GM/NIGMS NIH HHS/United States
- R01 CA233922/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

