A 3D Bioprinted Human Neurovascular Unit Model of Glioblastoma Tumor Growth
- PMID: 38394389
- PMCID: PMC11176035
- DOI: 10.1002/adhm.202302831
A 3D Bioprinted Human Neurovascular Unit Model of Glioblastoma Tumor Growth
Abstract
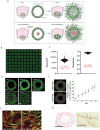
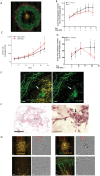
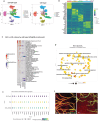
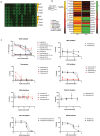
A 3D bioprinted neurovascular unit (NVU) model is developed to study glioblastoma (GBM) tumor growth in a brain-like microenvironment. The NVU model includes human primary astrocytes, pericytes and brain microvascular endothelial cells, and patient-derived glioblastoma cells (JHH-520) are used for this study. Fluorescence reporters are used with confocal high content imaging to quantitate real-time microvascular network formation and tumor growth. Extensive validation of the NVU-GBM model includes immunostaining for brain relevant cellular markers and extracellular matrix components; single cell RNA sequencing (scRNAseq) to establish physiologically relevant transcriptomics changes; and secretion of NVU and GBM-relevant cytokines. The scRNAseq reveals changes in gene expression and cytokines secretion associated with wound healing/angiogenesis, including the appearance of an endothelial mesenchymal transition cell population. The NVU-GBM model is used to test 18 chemotherapeutics and anti-cancer drugs to assess the pharmacological relevance of the model and robustness for high throughput screening.
Keywords: 3D Bioprinting; glioblastomas; high‐throughput screening; neurovascular unit; transcriptomics.
Published 2024. This article is a U.S. Government work and is in the public domain in the USA. Advanced Healthcare Materials published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

