Virtual histological staining of unlabeled autopsy tissue
- PMID: 38396004
- PMCID: PMC10891155
- DOI: 10.1038/s41467-024-46077-2
Virtual histological staining of unlabeled autopsy tissue
Abstract
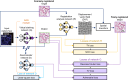
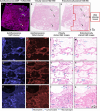
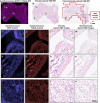
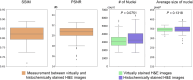
Traditional histochemical staining of post-mortem samples often confronts inferior staining quality due to autolysis caused by delayed fixation of cadaver tissue, and such chemical staining procedures covering large tissue areas demand substantial labor, cost and time. Here, we demonstrate virtual staining of autopsy tissue using a trained neural network to rapidly transform autofluorescence images of label-free autopsy tissue sections into brightfield equivalent images, matching hematoxylin and eosin (H&E) stained versions of the same samples. The trained model can effectively accentuate nuclear, cytoplasmic and extracellular features in new autopsy tissue samples that experienced severe autolysis, such as COVID-19 samples never seen before, where the traditional histochemical staining fails to provide consistent staining quality. This virtual autopsy staining technique provides a rapid and resource-efficient solution to generate artifact-free H&E stains despite severe autolysis and cell death, also reducing labor, cost and infrastructure requirements associated with the standard histochemical staining.
© 2024. The Author(s).
Conflict of interest statement
A.O. is the founder of a company (Pictor Labs) that commercializes virtual staining technology. A.O., K.H., J.L. and Y.Z. have pending patent applications on virtual tissue staining, related to the deep learning-based methods used in this manuscript on virtual histological staining of unlabeled autopsy tissue. The remaining authors declare no competing interests.
Figures



Similar articles
-
Use of Deep Learning to Develop and Analyze Computational Hematoxylin and Eosin Staining of Prostate Core Biopsy Images for Tumor Diagnosis.JAMA Netw Open. 2020 May 1;3(5):e205111. doi: 10.1001/jamanetworkopen.2020.5111. JAMA Netw Open. 2020. PMID: 32432709 Free PMC article.
-
A large-scale internal validation study of unsupervised virtual trichrome staining technologies on nonalcoholic steatohepatitis liver biopsies.Mod Pathol. 2021 Apr;34(4):808-822. doi: 10.1038/s41379-020-00718-1. Epub 2020 Dec 9. Mod Pathol. 2021. PMID: 33299110 Free PMC article.
-
Deep Learning for Virtual Histological Staining of Bright-Field Microscopic Images of Unlabeled Carotid Artery Tissue.Mol Imaging Biol. 2020 Oct;22(5):1301-1309. doi: 10.1007/s11307-020-01508-6. Mol Imaging Biol. 2020. PMID: 32514884 Free PMC article.
-
The hematoxylin and eosin stain in anatomic pathology-An often-neglected focus of quality assurance in the laboratory.Semin Diagn Pathol. 2019 Sep;36(5):303-311. doi: 10.1053/j.semdp.2019.06.003. Epub 2019 Jun 4. Semin Diagn Pathol. 2019. PMID: 31230963 Review.
-
Hematoxylin and eosin tissue stain in Mohs micrographic surgery: a review.Dermatol Surg. 2011 Aug;37(8):1089-99. doi: 10.1111/j.1524-4725.2011.02051.x. Epub 2011 Jun 2. Dermatol Surg. 2011. PMID: 21635628 Review.
Cited by
-
Virtual Gram staining of label-free bacteria using dark-field microscopy and deep learning.Sci Adv. 2025 Jan 10;11(2):eads2757. doi: 10.1126/sciadv.ads2757. Epub 2025 Jan 8. Sci Adv. 2025. PMID: 39772690 Free PMC article.
-
A Benchmark for Virus Infection Reporter Virtual Staining in Fluorescence and Brightfield Microscopy.Sci Data. 2025 May 28;12(1):886. doi: 10.1038/s41597-025-05194-3. Sci Data. 2025. PMID: 40436865 Free PMC article.
-
ML-driven segmentation of microvascular features during histological examination of tissue-engineered vascular grafts.Front Bioeng Biotechnol. 2024 Jun 26;12:1411680. doi: 10.3389/fbioe.2024.1411680. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 38988863 Free PMC article.
-
In vivo organoid growth monitoring by stimulated Raman histology.Npj Imaging. 2024;2(1):18. doi: 10.1038/s44303-024-00019-1. Epub 2024 Jun 28. Npj Imaging. 2024. PMID: 38948153 Free PMC article.
-
UniFORM: Towards Universal ImmunoFluorescence Normalization for Multiplex Tissue Imaging.bioRxiv [Preprint]. 2025 May 14:2024.12.06.626879. doi: 10.1101/2024.12.06.626879. bioRxiv. 2025. PMID: 39713407 Free PMC article. Preprint.
References
-
- Molina DK, Wood LE, Frost RE. Is Routine Histopathologic Examination Beneficial in All Medicolegal Autopsies? Am. J. Forensic Med. Pathol. 2007;28:1. - PubMed
-
- Khoury T. Delay to formalin fixation alters morphology and immunohistochemistry for breast carcinoma. Appl Immunohistochem. Mol. Morphol. 2012;20:531–542. - PubMed

