Randomized controlled trial of molnupiravir SARS-CoV-2 viral and antibody response in at-risk adult outpatients
- PMID: 38396069
- PMCID: PMC10891158
- DOI: 10.1038/s41467-024-45641-0
Randomized controlled trial of molnupiravir SARS-CoV-2 viral and antibody response in at-risk adult outpatients
Abstract
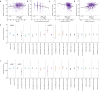
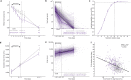
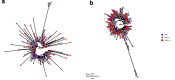
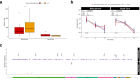
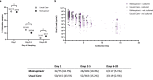
Viral clearance, antibody response and the mutagenic effect of molnupiravir has not been elucidated in at-risk populations. Non-hospitalised participants within 5 days of SARS-CoV-2 symptoms randomised to receive molnupiravir (n = 253) or Usual Care (n = 324) were recruited to study viral and antibody dynamics and the effect of molnupiravir on viral whole genome sequence from 1437 viral genomes. Molnupiravir accelerates viral load decline, but virus is detectable by Day 5 in most cases. At Day 14 (9 days post-treatment), molnupiravir is associated with significantly higher viral persistence and significantly lower anti-SARS-CoV-2 spike antibody titres compared to Usual Care. Serial sequencing reveals increased mutagenesis with molnupiravir treatment. Persistence of detectable viral RNA at Day 14 in the molnupiravir group is associated with higher transition mutations following treatment cessation. Viral viability at Day 14 is similar in both groups with post-molnupiravir treated samples cultured up to 9 days post cessation of treatment. The current 5-day molnupiravir course is too short. Longer courses should be tested to reduce the risk of potentially transmissible molnupiravir-mutated variants being generated. Trial registration: ISRCTN30448031.
© 2024. The Author(s).
Conflict of interest statement
J.F.S. has participated on a data safety monitoring board for GlaxoSmithKline (Sotrovimab) with fees paid to his institution. JSN-V-T was seconded to the Department of Health and Social Care, England (DHSC) from October 2017–March 2022. The views and opinions expressed in this paper are not necessarily those of DHSC or any of its arms-length bodies. JSN-V-T performed one-off paid consultancy for Merck Sharp and Dohme in June 2023, unrelated to the subject of the manuscript. K.H. was a member of the Health Technology Assessment General Committee and Funding Strategy Group until November 2022, and Research Professors Funding Committee at the UK National Institute for Health and Care Research (NIHR), received a grant from AstraZeneca (paid to their institution) to support a trial of Evusheld for the prevention of COVID-19 in high-risk individuals (RAPID-Protection), and was an independent member of the independent data monitoring committee for the OCTAVE-DUO trial of vaccines in individuals at high risk of COVID-19. D.M.L. has received grants or contracts from LifeArc, the UK Medical Research Council, Bristol Myers Squibb, GlaxoSmithKline, the British Society for Antimicrobial Chemotherapy, and Blood Cancer UK, personal fees or honoraria from Biotest UK, Gilead, and Merck, consulting fees from GlaxoSmithKline (paid to their institution), and conference support from Octapharma. DBR has received consulting fees from OMASS Therapeutics, GSK, and Sosei-Heptares and has a leadership and fiduciary role in the Heal-COVID trial TMG. M.L. is a member of the data monitoring and ethics committee of RAPIS-TEST (NIHR efficacy and mechanism evaluation). S.K. reports grants from GlaxoSmithKline, ViiV, Ridgeback Biotherapeutics, Vir, Merck, the UK Medical Research Council, and the Wellcome Trust (all paid to his institution), speaker’s honoraria from ViiV, and donations of drugs for clinical studies from ViiV Healthcare, Toyama, and GlaxoSmithKline. M.A. has received grants from the Blood and Transplant Research Unit, Janssen, Pfizer, Prenetics, Dunhill Medical Trust, the BMA Trust (Kathleen Harper Fund), and Antibiotic Research UK (all of which were paid to their institution), and consultancy fees from Prenetics and OxDx. M.A. reports a planned patent for Ramanomics, has participated on data safety monitoring boards or advisory boards for Prenetics, and has an unpaid leadership or fiduciary role in the E3 Initiative. NPBT has received payment for participation on an advisory board from MSD (before any knowledge or planning of this trial). O.v.H. has received consulting fees from MindGap (fees paid to Oxford University lnnovation), has participated on data safety monitoring boards or advisory boards for the CHICO trial, and has an unpaid leadership or fiduciary role in the British Society of Antimicrobial Chemotherapy. J.B. has received consulting fees from GlaxoSmithKline (paid to her institution). All other authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

