Multi-scale V-net architecture with deep feature CRF layers for brain extraction
- PMID: 38396078
- PMCID: PMC10891085
- DOI: 10.1038/s43856-024-00452-8
Multi-scale V-net architecture with deep feature CRF layers for brain extraction
Abstract
Background: Brain extraction is a computational necessity for researchers using brain imaging data. However, the complex structure of the interfaces between the brain, meninges and human skull have not allowed a highly robust solution to emerge. While previous methods have used machine learning with structural and geometric priors in mind, with the development of Deep Learning (DL), there has been an increase in Neural Network based methods. Most proposed DL models focus on improving the training data despite the clear gap between groups in the amount and quality of accessible training data between.
Methods: We propose an architecture we call Efficient V-net with Additional Conditional Random Field Layers (EVAC+). EVAC+ has 3 major characteristics: (1) a smart augmentation strategy that improves training efficiency, (2) a unique way of using a Conditional Random Fields Recurrent Layer that improves accuracy and (3) an additional loss function that fine-tunes the segmentation output. We compare our model to state-of-the-art non-DL and DL methods.
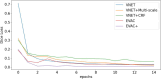
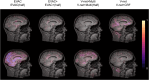
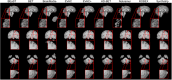
Results: Results show that even with limited training resources, EVAC+ outperforms in most cases, achieving a high and stable Dice Coefficient and Jaccard Index along with a desirable lower Surface (Hausdorff) Distance. More importantly, our approach accurately segmented clinical and pediatric data, despite the fact that the training dataset only contains healthy adults.
Conclusions: Ultimately, our model provides a reliable way of accurately reducing segmentation errors in complex multi-tissue interfacing areas of the brain. We expect our method, which is publicly available and open-source, to be beneficial to a wide range of researchers.
Plain language summary
Computational processing of brain images can enable better understanding and diagnosis of diseases that affect the brain. Brain Extraction is a computational method that can be used to remove areas of the head that are not the brain from images of the head. We compared various different computational methods that are available and used them to develop a better method. The method we describe in the paper is more accurate at imaging the brain of both healthy individuals and those known to have diseases that affect the brain than the other methods we evaluated. Our method might enable better understanding and diagnosis of diseases that affect the brain in the future.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Rehman HZU, Hwang H, Lee S. Conventional and deep learning methods for skull stripping in brain MRI. Appl. Sci. 2020;10:1773. doi: 10.3390/app10051773. - DOI
-
- Atkins, M. S., Siu, K., Law, B., Orchard, J. J. & Rosenbaum, W. L. Difficulties of t1 brain MRI segmentation techniques. In: Medical Imaging 2002: Image Processing (eds Milan, S., J & Michael, F.), 1837–1844 (SPIE, 2002).
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

