VSpipe-GUI, an Interactive Graphical User Interface for Virtual Screening and Hit Selection
- PMID: 38396680
- PMCID: PMC10889202
- DOI: 10.3390/ijms25042002
VSpipe-GUI, an Interactive Graphical User Interface for Virtual Screening and Hit Selection
Abstract
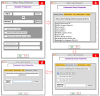
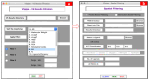
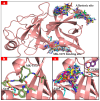
Virtual screening of large chemical libraries is essential to support computer-aided drug development, providing a rapid and low-cost approach for further experimental validation. However, existing computational packages are often for specialised users or platform limited. Previously, we developed VSpipe, an open-source semi-automated pipeline for structure-based virtual screening. We have now improved and expanded the initial command-line version into an interactive graphical user interface: VSpipe-GUI, a cross-platform open-source Python toolkit functional in various operating systems (e.g., Linux distributions, Windows, and Mac OS X). The new implementation is more user-friendly and accessible, and considerably faster than the previous version when AutoDock Vina is used for docking. Importantly, we have introduced a new compound selection module (i.e., spatial filtering) that allows filtering of docked compounds based on specified features at the target binding site. We have tested the new VSpipe-GUI on the Hepatitis C Virus NS3 (HCV NS3) protease as the target protein. The pocket-based and interaction-based modes of the spatial filtering module showed efficient and specific selection of ligands from the virtual screening that interact with the HCV NS3 catalytic serine 139.
Keywords: AutoDock; AutoDock Vina; VSpipe; VSpipe-GUI; chemoinformatics; docking; drug design; virtual screening.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



References
-
- Schrödinger L., DeLano W. PyMOL. 2020. [(accessed on 4 July 2023)]. Available online: http://www.pymol.org/pymol.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

