Impaired Mitochondrial Function and Marrow Failure in Patients Carrying a Variant of the SRSF4 Gene
- PMID: 38396760
- PMCID: PMC10888539
- DOI: 10.3390/ijms25042083
Impaired Mitochondrial Function and Marrow Failure in Patients Carrying a Variant of the SRSF4 Gene
Abstract
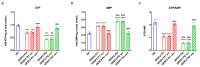
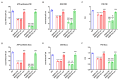
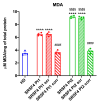
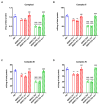
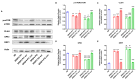
Serine/arginine-rich splicing factors (SRSFs) are a family of proteins involved in RNA metabolism, including pre-mRNA constitutive and alternative splicing. The role of SRSF proteins in regulating mitochondrial activity has already been shown for SRSF6, but SRSF4 altered expression has never been reported as a cause of bone marrow failure. An 8-year-old patient admitted to the hematology unit because of leukopenia, lymphopenia, and neutropenia showed a missense variant of unknown significance of the SRSF4 gene (p.R235W) found via whole genome sequencing analysis and inherited from the mother who suffered from mild leuko-neutropenia. Both patients showed lower SRSF4 protein expression and altered mitochondrial function and energetic metabolism in primary lymphocytes and Epstein-Barr-virus (EBV)-immortalized lymphoblasts compared to healthy donor (HD) cells, which appeared associated with low mTOR phosphorylation and an imbalance in the proteins regulating mitochondrial biogenesis (i.e., CLUH) and dynamics (i.e., DRP1 and OPA1). Transfection with the wtSRSF4 gene restored mitochondrial function. In conclusion, this study shows that the described variant of the SRSF4 gene is pathogenetic and causes reduced SRSF4 protein expression, which leads to mitochondrial dysfunction. Since mitochondrial function is crucial for hematopoietic stem cell maintenance and some genetic bone marrow failure syndromes display mitochondrial defects, the SRSF4 mutation could have substantially contributed to the clinical phenotype of our patient.
Keywords: CLUH; DRP1; OPA1; SRSF4; mTOR; marrow failure; mitochondria.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Fioredda F., Rotulo G.A., Farruggia P., Dagliano F., Pillon M., Trizzino A., Notarangelo L., Luti L., Lanza T., Terranova P., et al. Late-onset and long-lasting autoimmune neutropenia: An analysis from the Italian Neutropenia Registry. Blood Adv. 2020;4:5644–5649. doi: 10.1182/bloodadvances.2020002793. - DOI - PMC - PubMed
-
- Cifaldi C., Brigida I., Barzaghi F., Zoccolillo M., Ferradini V., Petricone D., Cicalese M.P., Lazarevic D., Cittaro D., Omrani M., et al. Targeted NGS Platforms for Genetic Screening and Gene Discovery in Primary Immunodeficiencies. Front. Immunol. 2019;10:316. doi: 10.3389/fimmu.2019.00316. - DOI - PMC - PubMed
-
- Kim E., Ilagan J.O., Liang Y., Daubner G.M., Lee S.C.W., Ramakrishnan A., Li Y., Chung Y.R., Micol J.-B., Murphy M.E., et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell. 2015;27:617–630. doi: 10.1016/j.ccell.2015.04.006. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

