The African Swine Fever Virus Virulence Determinant DP96R Suppresses Type I IFN Production Targeting IRF3
- PMID: 38396775
- PMCID: PMC10889005
- DOI: 10.3390/ijms25042099
The African Swine Fever Virus Virulence Determinant DP96R Suppresses Type I IFN Production Targeting IRF3
Abstract
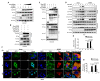
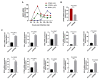
DP96R of African swine fever virus (ASFV), also known as uridine kinase (UK), encodes a virulence-associated protein. Previous studies have examined DP96R along with other genes in an effort to create live attenuated vaccines. While experiments in pigs have explored the impact of DP96R on the pathogenicity of ASFV, the precise molecular mechanism underlying this phenomenon remains unknown. Here, we describe a novel molecular mechanism by which DP96R suppresses interferon regulator factor-3 (IRF3)-mediated antiviral immune responses. DP96R interacts with a crucial karyopherin (KPNA) binding site within IRF3, disrupting the KPNA-IRF3 interaction and consequently impeding the translocation of IRF3 to the nucleus. Under this mechanistic basis, the ectopic expression of DP96R enhances the replication of DNA and RNA viruses by inhibiting the production of IFNs, whereas DP96R knock-down resulted in higher IFNs and IFN-stimulated gene (ISG) transcription during ASFV infection. Collectively, these findings underscore the pivotal role of DP96R in inhibiting IFN responses and increase our understanding of the relationship between DP96R and the virulence of ASFV.
Keywords: African swine fever virus; DP96R (UK gene); IRF3; KPNA.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
African Swine Fever Virus E120R Protein Inhibits Interferon Beta Production by Interacting with IRF3 To Block Its Activation.J Virol. 2021 Aug 25;95(18):e0082421. doi: 10.1128/JVI.00824-21. Epub 2021 Aug 25. J Virol. 2021. PMID: 34190598 Free PMC article.
-
African Swine Fever Virus Armenia/07 Virulent Strain Controls Interferon Beta Production through the cGAS-STING Pathway.J Virol. 2019 May 29;93(12):e02298-18. doi: 10.1128/JVI.02298-18. Print 2019 Jun 15. J Virol. 2019. PMID: 30918080 Free PMC article.
-
Inhibition of cGAS-STING-TBK1 signaling pathway by DP96R of ASFV China 2018/1.Biochem Biophys Res Commun. 2018 Nov 30;506(3):437-443. doi: 10.1016/j.bbrc.2018.10.103. Epub 2018 Oct 20. Biochem Biophys Res Commun. 2018. PMID: 30348523
-
Identification and utility of innate immune system evasion mechanisms of ASFV.Virus Res. 2013 Apr;173(1):87-100. doi: 10.1016/j.virusres.2012.10.013. Epub 2012 Nov 16. Virus Res. 2013. PMID: 23165138 Review.
-
African swine fever viral proteins that inhibit cGAS-STING pathway and type-I interferon production.Virology. 2025 Jan;602:110317. doi: 10.1016/j.virol.2024.110317. Epub 2024 Nov 26. Virology. 2025. PMID: 39616703 Review.
Cited by
-
The African swine fever virus B125R protein antagonizes JAK-STAT signalling by promoting the degradation of IFNAR2.Vet Res. 2025 Apr 23;56(1):87. doi: 10.1186/s13567-025-01523-x. Vet Res. 2025. PMID: 40270033 Free PMC article.
-
Advancement in the Antigenic Epitopes and Vaccine Adjuvants of African Swine Fever Virus.Pathogens. 2024 Aug 21;13(8):706. doi: 10.3390/pathogens13080706. Pathogens. 2024. PMID: 39204306 Free PMC article. Review.
-
Advances in African swine fever virus molecular biology and host interactions contributing to new tools for control.J Virol. 2025 Jun 17;99(6):e0093224. doi: 10.1128/jvi.00932-24. Epub 2025 May 9. J Virol. 2025. PMID: 40340396 Free PMC article. Review.
-
Double Deletion of EP402R and EP153R in the Attenuated Lv17/WB/Rie1 African Swine Fever Virus (ASFV) Enhances Safety, Provides DIVA Compatibility, and Confers Complete Protection Against a Genotype II Virulent Strain.Vaccines (Basel). 2024 Dec 13;12(12):1406. doi: 10.3390/vaccines12121406. Vaccines (Basel). 2024. PMID: 39772067 Free PMC article.
-
Inhibition of STING-mediated type I IFN signaling by African swine fever virus DP71L.Vet Res. 2025 Feb 4;56(1):27. doi: 10.1186/s13567-025-01474-3. Vet Res. 2025. PMID: 39905555 Free PMC article.
References
-
- Wilkinson P. The persistence of African swine fever in Africa and the Mediterranean. Prev. Vet. Med. 1984;2:71–82. doi: 10.1016/0167-5877(84)90050-3. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

