The Role of Mechanotransduction in Contact Inhibition of Locomotion and Proliferation
- PMID: 38396812
- PMCID: PMC10889191
- DOI: 10.3390/ijms25042135
The Role of Mechanotransduction in Contact Inhibition of Locomotion and Proliferation
Abstract
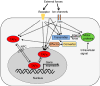
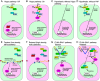
Contact inhibition (CI) represents a crucial tumor-suppressive mechanism responsible for controlling the unbridled growth of cells, thus preventing the formation of cancerous tissues. CI can be further categorized into two distinct yet interrelated components: CI of locomotion (CIL) and CI of proliferation (CIP). These two components of CI have historically been viewed as separate processes, but emerging research suggests that they may be regulated by both distinct and shared pathways. Specifically, recent studies have indicated that both CIP and CIL utilize mechanotransduction pathways, a process that involves cells sensing and responding to mechanical forces. This review article describes the role of mechanotransduction in CI, shedding light on how mechanical forces regulate CIL and CIP. Emphasis is placed on filamin A (FLNA)-mediated mechanotransduction, elucidating how FLNA senses mechanical forces and translates them into crucial biochemical signals that regulate cell locomotion and proliferation. In addition to FLNA, trans-acting factors (TAFs), which are proteins or regulatory RNAs capable of directly or indirectly binding to specific DNA sequences in distant genes to regulate gene expression, emerge as sensitive players in both the mechanotransduction and signaling pathways of CI. This article presents methods for identifying these TAF proteins and profiling the associated changes in chromatin structure, offering valuable insights into CI and other biological functions mediated by mechanotransduction. Finally, it addresses unanswered research questions in these fields and delineates their possible future directions.
Keywords: WW domain-containing transcription regulator protein 1 (TAZ); Yes-associated protein (YAP1); contact inhibition; contact inhibition of locomotion; contact inhibition of proliferation; filamin; gene expression; mechanical force; mechanotransduction; proteomics; ubiquitin-conjugating enzyme E2 A and B (UBE2A/B).
Conflict of interest statement
The authors declare no conflicts of interests.
Figures






Similar articles
-
UBE2A/B is the trans-acting factor mediating mechanotransduction and contact inhibition.Biochem J. 2023 Oct 31;480(20):1659-1674. doi: 10.1042/BCJ20230208. Biochem J. 2023. PMID: 37818922
-
Control of cellular responses to mechanical cues through YAP/TAZ regulation.J Biol Chem. 2019 Nov 15;294(46):17693-17706. doi: 10.1074/jbc.REV119.007963. Epub 2019 Oct 8. J Biol Chem. 2019. PMID: 31594864 Free PMC article. Review.
-
Cell response to mechanical microenvironment cues via Rho signaling: From mechanobiology to mechanomedicine.Acta Biomater. 2023 Mar 15;159:1-20. doi: 10.1016/j.actbio.2023.01.039. Epub 2023 Jan 28. Acta Biomater. 2023. PMID: 36717048 Review.
-
Contact inhibition (of proliferation) redux.Curr Opin Cell Biol. 2012 Oct;24(5):685-94. doi: 10.1016/j.ceb.2012.06.009. Epub 2012 Jul 24. Curr Opin Cell Biol. 2012. PMID: 22835462 Review.
-
Mechanosensing and mechanochemical transduction: how is mechanical energy sensed and converted into chemical energy in an extracellular matrix?Crit Rev Biomed Eng. 2003;31(4):255-331. doi: 10.1615/critrevbiomedeng.v31.i4.10. Crit Rev Biomed Eng. 2003. PMID: 15095950 Review.
Cited by
-
Comparative transcriptomic analysis validates iPSC derived in-vitro progressive fibrosis model as a screening tool for drug discovery and development in systemic sclerosis.Sci Rep. 2024 Oct 18;14(1):24428. doi: 10.1038/s41598-024-74610-2. Sci Rep. 2024. PMID: 39424619 Free PMC article.
-
Multicellular muscle-tendon bioprinting of mechanically optimized musculoskeletal bioactuators with enhanced force transmission.Sci Adv. 2025 Jul 18;11(29):eadv2628. doi: 10.1126/sciadv.adv2628. Epub 2025 Jul 16. Sci Adv. 2025. PMID: 40668913 Free PMC article.
-
Modulation of Gene Expression by Substrate Stiffness via Ubiquitination of Histone H2B by Ubiquitin-Conjugating Enzyme E2A/B.ACS Omega. 2025 Apr 9;10(15):15799-15809. doi: 10.1021/acsomega.5c02459. eCollection 2025 Apr 22. ACS Omega. 2025. PMID: 40290990 Free PMC article.
-
The WAVE complex forms linear arrays at negative membrane curvature to instruct lamellipodia formation.bioRxiv [Preprint]. 2025 Jun 15:2024.07.08.600855. doi: 10.1101/2024.07.08.600855. bioRxiv. 2025. Update in: J Cell Biol. 2025 Sep 1;224(9):e202410098. doi: 10.1083/jcb.202410098. PMID: 39026726 Free PMC article. Updated. Preprint.
-
The nucleocytoplasmic translocation of HINT1 regulates the maturation of cell density.Life Sci Alliance. 2025 Jul 22;8(10):e202503215. doi: 10.26508/lsa.202503215. Print 2025 Oct. Life Sci Alliance. 2025. PMID: 40695554 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

