Comparative Analysis of Chloroplast Genome of Meconopsis (Papaveraceae) Provides Insights into Their Genomic Evolution and Adaptation to High Elevation
- PMID: 38396871
- PMCID: PMC10888623
- DOI: 10.3390/ijms25042193
Comparative Analysis of Chloroplast Genome of Meconopsis (Papaveraceae) Provides Insights into Their Genomic Evolution and Adaptation to High Elevation
Abstract
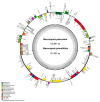
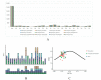
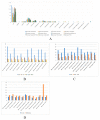
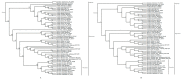
The Meconopsis species are widely distributed in the Qinghai-Tibet Plateau, Himalayas, and Hengduan Mountains in China, and have high medicinal and ornamental value. The high diversity of plant morphology in this genus poses significant challenges for species identification, given their propensity for highland dwelling, which makes it a question worth exploring how they cope with the harsh surroundings. In this study, we recently generated chloroplast (cp) genomes of two Meconopsis species, Meconopsis paniculata (M. paniculata) and M. pinnatifolia, and compared them with those of ten Meconopsis cp genomes to comprehend cp genomic features, their phylogenetic relationships, and what part they might play in plateau adaptation. These cp genomes shared a great deal of similarities in terms of genome size, structure, gene content, GC content, and codon usage patterns. The cp genomes were between 151,864 bp and 154,997 bp in length, and contain 133 predictive genes. Through sequence divergence analysis, we identified three highly variable regions (trnD-psbD, ccsA-ndhD, and ycf1 genes), which could be used as potential markers or DNA barcodes for phylogenetic analysis. Between 22 and 38 SSRs and some long repeat sequences were identified from 12 Meconopsis species. Our phylogenetic analysis confirmed that 12 species of Meconopsis clustered into a monophyletic clade in Papaveraceae, which corroborated their intrageneric relationships. The results indicated that M. pinnatifolia and M. paniculata are sister species in the phylogenetic tree. In addition, the atpA and ycf2 genes were positively selected in high-altitude species. The functions of these two genes might be involved in adaptation to the extreme environment in the cold and low CO2 concentration conditions at the plateau.
Keywords: Meconopsis; chloroplast genome; high altitude adaptation; high variation region.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Meger J., Ulaszewski B., Vendramin G.G., Burczyk J. Using reduced representation libraries sequencing methods to identify cpDNA polymorphisms in European beech (Fagus sylvatica L) Tree Genet. Genomes. 2019;15:7. doi: 10.1007/s11295-018-1313-6. - DOI
-
- Xiao W., Simpson B.B. A New Infrageneric Classification of Meconopsis (Papaveraceae) Based on a Well-supported Molecular Phylogeny. Syst. Bot. 2017;42:226–233. doi: 10.1600/036364417X695466. - DOI
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

