Genetic Biomarkers of Sorafenib Response in Patients with Hepatocellular Carcinoma
- PMID: 38396873
- PMCID: PMC10888718
- DOI: 10.3390/ijms25042197
Genetic Biomarkers of Sorafenib Response in Patients with Hepatocellular Carcinoma
Abstract
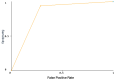
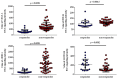
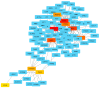
The identification of biomarkers for predicting inter-individual sorafenib response variability could allow hepatocellular carcinoma (HCC) patient stratification. SNPs in angiogenesis- and drug absorption, distribution, metabolism, and excretion (ADME)-related genes were evaluated to identify new potential predictive biomarkers of sorafenib response in HCC patients. Five known SNPs in angiogenesis-related genes, including VEGF-A, VEGF-C, HIF-1a, ANGPT2, and NOS3, were investigated in 34 HCC patients (9 sorafenib responders and 25 non-responders). A subgroup of 23 patients was genotyped for SNPs in ADME genes. A machine learning classifier method was used to discover classification rules for our dataset. We found that only the VEGF-A (rs2010963) C allele and CC genotype were significantly associated with sorafenib response. ADME-related gene analysis identified 10 polymorphic variants in ADH1A (rs6811453), ADH6 (rs10008281), SULT1A2/CCDC101 (rs11401), CYP26A1 (rs7905939), DPYD (rs2297595 and rs1801265), FMO2 (rs2020863), and SLC22A14 (rs149738, rs171248, and rs183574) significantly associated with sorafenib response. We have identified a genetic signature of predictive response that could permit non-responder/responder patient stratification. Angiogenesis- and ADME-related genes correlation was confirmed by cumulative genetic risk score and network and pathway enrichment analysis. Our findings provide a proof of concept that needs further validation in follow-up studies for HCC patient stratification for sorafenib prescription.
Keywords: anticancer drugs; genetic polymorphism; genotyping; pharmacogenomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

