Genomic and Immunologic Correlates in Prostate Cancer with High Expression of KLK2
- PMID: 38396898
- PMCID: PMC10889228
- DOI: 10.3390/ijms25042222
Genomic and Immunologic Correlates in Prostate Cancer with High Expression of KLK2
Abstract
The identification of surfaceome proteins is a main goal in cancer research to design antibody-based therapeutic strategies. T cell engagers based on KLK2, a kallikrein specifically expressed in prostate cancer (PRAD), are currently in early clinical development. Using genomic information from different sources, we evaluated the immune microenvironment and genomic profile of prostate tumors with high expression of KLK2. KLK2 was specifically expressed in PRAD but it was not significant associated with Gleason score. Additionally, KLK2 expression did not associate with the presence of any immune cell population and T cell activating markers. A mild correlation between the high expression of KLK2 and the deletion of TMPRSS2 was identified. KLK2 expression associated with high levels of surface proteins linked with a detrimental response to immune checkpoint inhibitors (ICIs) including CHRNA2, FAM174B, OR51E2, TSPAN1, PTPRN2, and the non-surface protein TRPM4. However, no association of these genes with an outcome in PRAD was observed. Finally, the expression of these genes in PRAD did not associate with an outcome in PRAD and any immune populations. We describe the immunologic microenvironment on PRAD tumors with a high expression of KLK2, including a gene signature linked with an inert immune microenvironment, that predicts the response to ICIs in other tumor types. Strategies targeting KLK2 with T cell engagers or antibody-drug conjugates will define whether T cell mobilization or antigen release and stimulation of immune cell death are sufficient effects to induce clinical activity.
Keywords: KLK2; T cell engagers; immunologic profile; prostate cancer; surfaceome.
Conflict of interest statement
A.O. is a former employee of Symphogen, Copengahen, Denmark and a current consultant of NMS. There are no conflicts of interest to declare in relation to this work. I.M.reports an advisory role in ESAME, Exafield and Guidepoint outside the submitted work. A.M. reports a consulting role in Grunenthal, outside the submitted work. V.M. reports personal fees from Bristol-Myers Squibb, Bayer, Janssen, and Pieris outside the submitted work. E.C. reports grants and personal fees from Astellas, Novartis, Nanobiotix, Pfizer, Janssen-Cilag, PsiOxus Therapeutics, Merck, Bristol-Myers Squibb, Seattle Genetics, Boehringer Ingelheim, AstraZeneca, Roche/Genentech, Servier, Celgene, AbbVie, Amcure, Alkermes, PharmaMar, and BeiGene, outside the submitted work. P.P. reports grant/research support from Bristol-Myers Squibb, AstraZeneca, and MSD, outside the submitted work. The other authors declare that they have no competing interests.
Figures



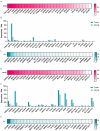
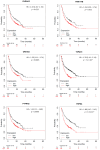
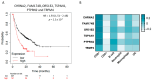
References
-
- Rudolph J., Settleman J., Malek S. Emerging Trends in Cancer Drug Discovery-From Drugging the “Undruggable” to Overcoming Resistance. Cancer Discov. 2021;11:815–821. doi: 10.1158/2159-8290.CD-21-0260. - DOI - PubMed
-
- Nieto-Jimenez C., Sanvicente A., Diaz-Tejeiro C., Moreno V., Lopez de Sa A., Calvo E., Martinez-Lopez J., Perez-Segura P., Ocana A. Uncovering therapeutic opportunities in the clinical development of antibody-drug conjugates. Clin. Transl. Med. 2023;13:e1329. doi: 10.1002/ctm2.1329. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

