Emerging Gene Therapeutics for Epidermolysis Bullosa under Development
- PMID: 38396920
- PMCID: PMC10889532
- DOI: 10.3390/ijms25042243
Emerging Gene Therapeutics for Epidermolysis Bullosa under Development
Abstract
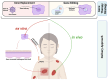
The monogenetic disease epidermolysis bullosa (EB) is characterised by the formation of extended blisters and lesions on the patient's skin upon minimal mechanical stress. Causal for this severe condition are genetic mutations in genes, leading to the functional impairment, reduction, or absence of the encoded protein within the skin's basement membrane zone connecting the epidermis to the underlying dermis. The major burden of affected families justifies the development of long-lasting and curative therapies operating at the genomic level. The landscape of causal therapies for EB is steadily expanding due to recent breakthroughs in the gene therapy field, providing promising outcomes for patients suffering from this severe disease. Currently, two gene therapeutic approaches show promise for EB. The clinically more advanced gene replacement strategy was successfully applied in severe EB forms, leading to a ground-breaking in vivo gene therapy product named beremagene geperpavec (B-VEC) recently approved from the US Food and Drug Administration (FDA). In addition, the continuous innovations in both designer nucleases and gene editing technologies enable the efficient and potentially safe repair of mutations in EB in a potentially permanent manner, inspiring researchers in the field to define and reach new milestones in the therapy of EB.
Keywords: base editing; epidermolysis bullosa; gene editing; gene replacement; gene therapy; prime editing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Has C., Bauer J.W., Bodemer C., Bolling M.C., Bruckner-Tuderman L., Diem A., Fine J.-D., Heagerty A., Hovnanian A., Marinkovich M.P., et al. Consensus reclassification of inherited epidermolysis bullosa and other disorders with skin fragility. Br. J. Dermatol. 2020;183:614–627. doi: 10.1111/bjd.18921. - DOI - PubMed
-
- Petrof G., Papanikolaou M., Martinez A.E., Mellerio J.E., McGrath J.A., Bardhan A., Harper N., Heagerty A., Ogboli M., Chiswell C., et al. The epidemiology of epidermolysis bullosa in England and Wales: Data from the national epidermolysis bullosa database. Br. J. Dermatol. 2022;186:843–848. doi: 10.1111/bjd.20958. - DOI - PubMed
-
- Baardman R., Yenamandra V.K., Duipmans J.C., Pasmooij A.M.G., Jonkman M.F., van den Akker P.C., Bolling M.C. Novel insights into the epidemiology of epidermolysis bullosa (EB) from the Dutch EB Registry: EB more common than previously assumed? J. Eur. Acad. Dermatol. Venereol. 2021;35:995–1006. doi: 10.1111/jdv.17012. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

