Lysine Methylation-Dependent Proteolysis by the Malignant Brain Tumor (MBT) Domain Proteins
- PMID: 38396925
- PMCID: PMC10889763
- DOI: 10.3390/ijms25042248
Lysine Methylation-Dependent Proteolysis by the Malignant Brain Tumor (MBT) Domain Proteins
Abstract
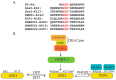
Lysine methylation is a major post-translational protein modification that occurs in both histones and non-histone proteins. Emerging studies show that the methylated lysine residues in non-histone proteins provide a proteolytic signal for ubiquitin-dependent proteolysis. The SET7 (SETD7) methyltransferase specifically transfers a methyl group from S-Adenosyl methionine to a specific lysine residue located in a methylation degron motif of a protein substrate to mark the methylated protein for ubiquitin-dependent proteolysis. LSD1 (Kdm1a) serves as a demethylase to dynamically remove the methyl group from the modified protein. The methylated lysine residue is specifically recognized by L3MBTL3, a methyl-lysine reader that contains the malignant brain tumor domain, to target the methylated proteins for proteolysis by the CRL4DCAF5 ubiquitin ligase complex. The methylated lysine residues are also recognized by PHF20L1 to protect the methylated proteins from proteolysis. The lysine methylation-mediated proteolysis regulates embryonic development, maintains pluripotency and self-renewal of embryonic stem cells and other stem cells such as neural stem cells and hematopoietic stem cells, and controls other biological processes. Dysregulation of the lysine methylation-dependent proteolysis is associated with various diseases, including cancers. Characterization of lysine methylation should reveal novel insights into how development and related diseases are regulated.
Keywords: CRL4; DCAF5; L3MBTL3; LSD1/Kdm1a; SET7/SETD7; demethylation; lysine methylation; ubiquitin E3 ligase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

