Engineered Chimera Protein Constructs to Facilitate the Production of Heterologous Transmembrane Proteins in E. coli
- PMID: 38397029
- PMCID: PMC10889703
- DOI: 10.3390/ijms25042354
Engineered Chimera Protein Constructs to Facilitate the Production of Heterologous Transmembrane Proteins in E. coli
Abstract
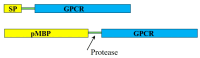
To delve into the structure-function relationship of transmembrane proteins (TMPs), robust protocols are needed to produce them in a pure, stable, and functional state. Among all hosts that express heterologous TMPs, E. coli has the lowest cost and fastest turnover. However, many of the TMPs expressed in E. coli are misfolded. Several strategies have been developed to either direct the foreign TMPs to E. coli's membrane or retain them in a cytosolic soluble form to overcome this deficiency. Here, we summarize protein engineering methods to produce chimera constructs of the desired TMPs fused to either a signal peptide or precursor maltose binding protein (pMBP) to direct the entire construct to the periplasm, therefore depositing the fused TMP in the plasma membrane. We further describe strategies to produce TMPs in soluble form by utilizing N-terminally fused MBP without a signal peptide. Depending on its N- or C-terminus location, a fusion to apolipoprotein AI can either direct the TMP to the membrane or shield the hydrophobic regions of the TMP, maintaining the soluble form. Strategies to produce G-protein-coupled receptors, TMPs of Mycobacterium tuberculosis, HIV-1 Vpu, and other TMPs are discussed. This knowledge could increase the scope of TMPs' expression in E. coli.
Keywords: E. coli expression host of heterologous transmembrane proteins; protein engineering; soluble transmembrane proteins; transmembrane protein fusion strategies.
Conflict of interest statement
The authors declare no conflict of interests.
Figures

Similar articles
-
On the mode of integration of the thylakoid membrane protein cytochrome b(6) into cytoplasmic membrane of Escherichia coli.Acta Biochim Pol. 2011;58(3):335-43. Epub 2011 Jul 5. Acta Biochim Pol. 2011. PMID: 21725502
-
Export of unprocessed precursor maltose-binding protein to the periplasm of Escherichia coli cells.J Bacteriol. 1987 Jun;169(6):2352-9. doi: 10.1128/jb.169.6.2352-2359.1987. J Bacteriol. 1987. PMID: 3294787 Free PMC article.
-
Export of maltose-binding protein species with altered charge distribution surrounding the signal peptide hydrophobic core in Escherichia coli cells harboring prl suppressor mutations.J Bacteriol. 1992 Jan;174(1):92-101. doi: 10.1128/jb.174.1.92-101.1992. J Bacteriol. 1992. PMID: 1729228 Free PMC article.
-
Enhancing the solubility of recombinant proteins in Escherichia coli by using hexahistidine-tagged maltose-binding protein as a fusion partner.Methods Mol Biol. 2011;705:259-74. doi: 10.1007/978-1-61737-967-3_16. Methods Mol Biol. 2011. PMID: 21125392 Review.
-
Transmembrane proteins in grape immunity: current knowledge and methodological advances.Front Plant Sci. 2024 Dec 20;15:1515163. doi: 10.3389/fpls.2024.1515163. eCollection 2024. Front Plant Sci. 2024. PMID: 39759230 Free PMC article. Review.
References
-
- Cournia Z., Allen T.W., Andricioaei I., Antonny B., Baum D., Brannigan G., Buchete N.-V., Deckman J.T., Delemotte L., Del Val C., et al. Membrane Protein Structure, Function, and Dynamics: A Perspective from Experiments and Theory. J. Membr. Biol. 2015;248:611–640. doi: 10.1007/s00232-015-9802-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

