Immunoinformatic Identification of Multiple Epitopes of gp120 Protein of HIV-1 to Enhance the Immune Response against HIV-1 Infection
- PMID: 38397105
- PMCID: PMC10889372
- DOI: 10.3390/ijms25042432
Immunoinformatic Identification of Multiple Epitopes of gp120 Protein of HIV-1 to Enhance the Immune Response against HIV-1 Infection
Abstract
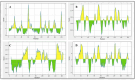
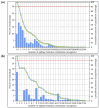
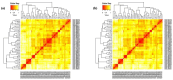
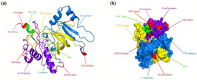
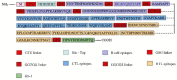
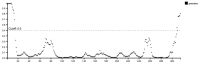
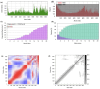
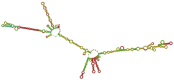
Acquired Immunodeficiency Syndrome is caused by the Human Immunodeficiency Virus (HIV), and a significant number of fatalities occur annually. There is a dire need to develop an effective vaccine against HIV-1. Understanding the structural proteins of viruses helps in designing a vaccine based on immunogenic peptides. In the current experiment, we identified gp120 epitopes using bioinformatic epitope prediction tools, molecular docking, and MD simulations. The Gb-1 peptide was considered an adjuvant. Consecutive sequences of GTG, GSG, GGTGG, and GGGGS linkers were used to bind the B cell, Cytotoxic T Lymphocytes (CTL), and Helper T Lymphocytes (HTL) epitopes. The final vaccine construct consisted of 315 amino acids and is expected to be a recombinant protein of approximately 35.49 kDa. Based on docking experiments, molecular dynamics simulations, and tertiary structure validation, the analysis of the modeled protein indicates that it possesses a stable structure and can interact with Toll-like receptors. The analysis demonstrates that the proposed vaccine can provoke an immunological response by activating T and B cells, as well as stimulating the release of IgA and IgG antibodies. This vaccine shows potential for HIV-1 prophylaxis. The in-silico design suggests that multiple-epitope constructs can be used as potentially effective immunogens for HIV-1 vaccine development.
Keywords: HIV-1; epitope; gp120; in silico.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures








Similar articles
-
Immunoinformatic-driven design and evaluation of multi-epitope mRNA vaccine targeting HIV-1 gp120.Front Immunol. 2025 May 13;16:1480025. doi: 10.3389/fimmu.2025.1480025. eCollection 2025. Front Immunol. 2025. PMID: 40433366 Free PMC article.
-
Exploring T & B-cell epitopes and designing multi-epitope subunit vaccine targeting integration step of HIV-1 lifecycle using immunoinformatics approach.Microb Pathog. 2019 Dec;137:103791. doi: 10.1016/j.micpath.2019.103791. Epub 2019 Oct 10. Microb Pathog. 2019. PMID: 31606417
-
Prioritization of potential vaccine candidates and designing a multiepitope-based subunit vaccine against multidrug-resistant Salmonella Typhi str. CT18: A subtractive proteomics and immunoinformatics approach.Microb Pathog. 2021 Oct;159:105150. doi: 10.1016/j.micpath.2021.105150. Epub 2021 Aug 20. Microb Pathog. 2021. PMID: 34425197
-
Epitope Prediction by Novel Immunoinformatics Approach: A State-of-the-art Review.Int J Pept Res Ther. 2020;26(2):1155-1163. doi: 10.1007/s10989-019-09918-z. Epub 2019 Aug 20. Int J Pept Res Ther. 2020. PMID: 32435171 Free PMC article. Review.
-
Fungal Vaccine Development: State of the Art and Perspectives Using Immunoinformatics.J Fungi (Basel). 2023 May 31;9(6):633. doi: 10.3390/jof9060633. J Fungi (Basel). 2023. PMID: 37367569 Free PMC article. Review.
Cited by
-
In Silico Subtractive Proteome Analysis to Design Multi-Epitope-Based Subunit Vaccine against Eikenella corrodens.J Microbiol Biotechnol. 2024 Nov 25;35:e2410015. doi: 10.4014/jmb.2410.10015. J Microbiol Biotechnol. 2024. PMID: 39809513 Free PMC article.
-
An integrated mutation-based immunoinformatic approach incorporating variability in epitopes: a study based on HIV subtype C.Front Immunol. 2025 May 20;16:1540253. doi: 10.3389/fimmu.2025.1540253. eCollection 2025. Front Immunol. 2025. PMID: 40463364 Free PMC article.
-
An mRNA vaccine candidate encoding cholera toxin subunit B and conserved antigens of influenza viruses confers cross-protection against influenza a viruses in adult and aged mice.Hum Vaccin Immunother. 2025 Dec;21(1):2453304. doi: 10.1080/21645515.2025.2453304. Epub 2025 Feb 16. Hum Vaccin Immunother. 2025. PMID: 39957235 Free PMC article.
-
Exploratory algorithms to devise multi-epitope subunit vaccine by examining HIV-1 envelope glycoprotein: An immunoinformatics and viroinformatics approach.PLoS One. 2025 Feb 27;20(2):e0318523. doi: 10.1371/journal.pone.0318523. eCollection 2025. PLoS One. 2025. Retraction in: PLoS One. 2025 May 6;20(5):e0324076. doi: 10.1371/journal.pone.0324076. PMID: 40014623 Free PMC article. Retracted.
-
Designing a multi-epitope vaccine against African swine fever virus using immunoinformatics approach.Sci Rep. 2025 May 8;15(1):16044. doi: 10.1038/s41598-025-00705-z. Sci Rep. 2025. PMID: 40341420 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

