Genome-Wide Classification of Myb Domain-Containing Protein Families in Entamoeba invadens
- PMID: 38397191
- PMCID: PMC10887745
- DOI: 10.3390/genes15020201
Genome-Wide Classification of Myb Domain-Containing Protein Families in Entamoeba invadens
Abstract
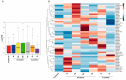
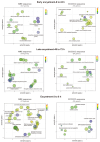
Entamoeba histolytica, the causative agent of amebiasis, is the third leading cause of death among parasitic diseases globally. Its life cycle includes encystation, which has been mostly studied in Entamoeba invadens, responsible for reptilian amebiasis. However, the molecular mechanisms underlying this process are not fully understood. Therefore, we focused on the identification and characterization of Myb proteins, which regulate the expression of encystation-related genes in various protozoan parasites. Through bioinformatic analysis, we identified 48 genes in E. invadens encoding MYB-domain-containing proteins. These were classified into single-repeat 1R (20), 2R-MYB proteins (27), and one 4R-MYB protein. The in-silico analysis suggests that these proteins are multifunctional, participating in transcriptional regulation, chromatin remodeling, telomere maintenance, and splicing. Transcriptomic data analysis revealed expression signatures of eimyb genes, suggesting a potential orchestration in the regulation of early and late encystation-excystation genes. Furthermore, we identified probable target genes associated with reproduction, the meiotic cell cycle, ubiquitin-dependent protein catabolism, and endosomal transport. In conclusion, our findings suggest that E. invadens Myb proteins regulate stage-specific proteins and a wide array of cellular processes. This study provides a foundation for further exploration of the molecular mechanisms governing encystation and unveils potential targets for therapeutic intervention in amebiasis.
Keywords: MYB-DBD-containing proteins; Myb recognition element; encystation–excystation; protozoan; transcriptional regulation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Transcriptome analysis of encystation in Entamoeba invadens.PLoS One. 2013 Sep 11;8(9):e74840. doi: 10.1371/journal.pone.0074840. eCollection 2013. PLoS One. 2013. PMID: 24040350 Free PMC article.
-
Expression analysis of Entamoeba invadens profilins in encystation and excystation.Parasitol Res. 2012 Jun;110(6):2095-104. doi: 10.1007/s00436-011-2735-3. Epub 2011 Dec 17. Parasitol Res. 2012. PMID: 22179263
-
Different structure and mRNA expression of Entamoeba invadens chitinases in the encystation and excystation.Parasitol Res. 2011 Aug;109(2):417-23. doi: 10.1007/s00436-011-2270-2. Epub 2011 Feb 1. Parasitol Res. 2011. PMID: 21286750
-
Entamoeba Encystation: New Targets to Prevent the Transmission of Amebiasis.PLoS Pathog. 2016 Oct 20;12(10):e1005845. doi: 10.1371/journal.ppat.1005845. eCollection 2016 Oct. PLoS Pathog. 2016. PMID: 27764256 Free PMC article. Review.
-
Conservation and function of Rab small GTPases in Entamoeba: annotation of E. invadens Rab and its use for the understanding of Entamoeba biology.Exp Parasitol. 2010 Nov;126(3):337-47. doi: 10.1016/j.exppara.2010.04.014. Epub 2010 Apr 29. Exp Parasitol. 2010. PMID: 20434444 Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

