Genome-Wide Detection for Runs of Homozygosity in Baoshan Pigs Using Whole Genome Resequencing
- PMID: 38397222
- PMCID: PMC10887577
- DOI: 10.3390/genes15020233
Genome-Wide Detection for Runs of Homozygosity in Baoshan Pigs Using Whole Genome Resequencing
Abstract
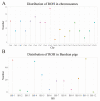
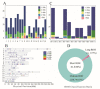
Baoshan pigs (BS) are a local breed in Yunnan Province that may face inbreeding owing to its limited population size. To accurately evaluate the inbreeding level of the BS pig population, we used whole-genome resequencing to identify runs of homozygosity (ROH) regions in BS pigs, calculated the inbreeding coefficient based on pedigree and ROH, and screened candidate genes with important economic traits from ROH islands. A total of 22,633,391 SNPS were obtained from the whole genome of BS pigs, and 201 ROHs were detected from 532,450 SNPS after quality control. The number of medium-length ROH (1-5 Mb) was the highest (98.43%), the number of long ROH (>5 Mb) was the lowest (1.57%), and the inbreeding of BS pigs mainly occurred in distant generations. The inbreeding coefficient FROH, calculated based on ROH, was 0.018 ± 0.016, and the FPED, calculated based on the pedigree, was 0.027 ± 0.028, which were positively correlated. Forty ROH islands were identified, containing 507 genes and 891 QTLs. Several genes were associated with growth and development (IGFALS, PTN, DLX5, DKK1, WNT2), meat quality traits (MC3R, ACSM3, ECI1, CD36, ROCK1, CACNA2D1), and reproductive traits (NPW, TSHR, BMP7). This study provides a reference for the protection and utilization of BS pigs.
Keywords: candidate gene; inbreeding coefficient; local breed; runs of homozygosity; whole genome resequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Genome-wide scan for runs of homozygosity in Asian wild boars and Anqing six-end-white pigs.Anim Genet. 2022 Dec;53(6):867-871. doi: 10.1111/age.13250. Epub 2022 Sep 9. Anim Genet. 2022. PMID: 36086874
-
Comparative evaluation of genomic inbreeding parameters in seven commercial and autochthonous pig breeds.Animal. 2020 May;14(5):910-920. doi: 10.1017/S175173111900332X. Epub 2020 Jan 13. Animal. 2020. PMID: 31928538
-
Assessment of runs of homozygosity islands and estimates of genomic inbreeding in Gyr (Bos indicus) dairy cattle.BMC Genomics. 2018 Jan 9;19(1):34. doi: 10.1186/s12864-017-4365-3. BMC Genomics. 2018. PMID: 29316879 Free PMC article.
-
Runs of homozygosity: current knowledge and applications in livestock.Anim Genet. 2017 Jun;48(3):255-271. doi: 10.1111/age.12526. Epub 2016 Dec 1. Anim Genet. 2017. PMID: 27910110 Review.
-
Genome-wide selection signatures detection in Shanghai Holstein cattle population identified genes related to adaption, health and reproduction traits.BMC Genomics. 2021 Oct 15;22(1):747. doi: 10.1186/s12864-021-08042-x. BMC Genomics. 2021. PMID: 34654366 Free PMC article. Review.
Cited by
-
Long-read and short-read RNA-seq reveal the transcriptional regulation characteristics of PICK1 in Baoshan pig testis.Anim Reprod. 2024 Oct 4;21(4):e20240047. doi: 10.1590/1984-3143-AR2024-0047. eCollection 2024. Anim Reprod. 2024. PMID: 39371543 Free PMC article.
References
-
- Chen H., Huang T., Zhang Z., Yang B., Jiang C., Wu J., Zhou Z., Zheng H., Xin W., Huang M., et al. Genome-wide association studies and meta-analysis reveal novel quantitative trait loci and pleiotropic loci for swine head-related traits. J. Anim. Sci. 2017;95:2354–2366. doi: 10.2527/jas2016.1137. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

