Satellitome Analysis in the Southern Lapwing (Vanellus chilensis) Genome: Implications for SatDNA Evolution in Charadriiform Birds
- PMID: 38397247
- PMCID: PMC10887557
- DOI: 10.3390/genes15020258
Satellitome Analysis in the Southern Lapwing (Vanellus chilensis) Genome: Implications for SatDNA Evolution in Charadriiform Birds
Abstract
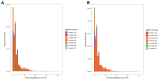
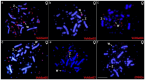
Vanellus (Charadriidae; Charadriiformes) comprises around 20 species commonly referred to as lapwings. In this study, by integrating cytogenetic and genomic approaches, we assessed the satellite DNA (satDNA) composition of one typical species, Vanellus chilensis, with a highly conserved karyotype. We additionally underlined its role in the evolution, structure, and differentiation process of the present ZW sex chromosome system. Seven distinct satellite DNA families were identified within its genome, accumulating on the centromeres, microchromosomes, and the W chromosome. However, these identified satellite DNA families were not found in two other Charadriiformes members, namely Jacana jacana and Calidris canutus. The hybridization of microsatellite sequences revealed the presence of a few repetitive sequences in V. chilensis, with only two out of sixteen displaying positive hybridization signals. Overall, our results contribute to understanding the genomic organization and satDNA evolution in Charadriiform birds.
Keywords: Charadriidae; chromosome evolution; constitutive heterochromatin; repetitive DNA; sex chromosomes.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Nie W., O’Brien P.C.M., Ng B.L., Fu B., Volobouev V., Carter N.P., Ferguson-Smith M.A., Yang F. Avian comparative genomics: Reciprocal chromosome painting between domestic chicken (Gallus gallus) and the stone curlew (Burhinus oedicnemus, Charadriiformes)—An atypical species with low diploid number. Chromosome Res. 2009;17:99–113. doi: 10.1007/s10577-009-9021-6. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

