Antioxidative Sirt1 and the Keap1-Nrf2 Signaling Pathway Impair Inflammation and Positively Regulate Autophagy in Murine Mammary Epithelial Cells or Mammary Glands Infected with Streptococcus uberis
- PMID: 38397769
- PMCID: PMC10886112
- DOI: 10.3390/antiox13020171
Antioxidative Sirt1 and the Keap1-Nrf2 Signaling Pathway Impair Inflammation and Positively Regulate Autophagy in Murine Mammary Epithelial Cells or Mammary Glands Infected with Streptococcus uberis
Abstract
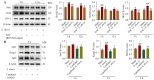
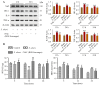
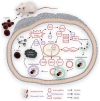
Streptococcus uberis mastitis in cattle infects mammary epithelial cells. Although oxidative responses often remove intracellular microbes, S. uberis survives, but the mechanisms are not well understood. Herein, we aimed to elucidate antioxidative mechanisms during pathogenesis of S. uberis after isolation from clinical bovine mastitis milk samples. S. uberis's in vitro pathomorphology, oxidative stress biological activities, transcription of antioxidative factors, inflammatory response cytokines, autophagosome and autophagy functions were evaluated, and in vivo S. uberis was injected into the fourth mammary gland nipple of each mouse to assess the infectiousness of S. uberis potential molecular mechanisms. The results showed that infection with S. uberis induced early oxidative stress and increased reactive oxygen species (ROS). However, over time, ROS concentrations decreased due to increased antioxidative activity, including total superoxide dismutase (T-SOD) and malondialdehyde (MDA) enzymes, plus transcription of antioxidative factors (Sirt1, Keap1, Nrf2, HO-1). Treatment with a ROS scavenger (N-acetyl cysteine, NAC) before infection with S. uberis reduced antioxidative responses and the inflammatory response, including the cytokines IL-6 and TNF-α, and the formation of the Atg5-LC3II/LC3I autophagosome. Synthesis of antioxidants determined autophagy functions, with Sirt1/Nrf2 activating autophagy in the presence of S. uberis. This study demonstrated the evasive mechanisms of S. uberis in mastitis, including suppressing inflammatory and ROS defenses by stimulating antioxidative pathways.
Keywords: Streptococcus uberis; antioxidative pathway; autophagy; bovine mastitis; inflammation; mMECs; murine mammary glands.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
Resveratrol alleviates oxidative stress caused by Streptococcus uberis infection via activating the Nrf2 signaling pathway.Int Immunopharmacol. 2020 Dec;89(Pt A):107076. doi: 10.1016/j.intimp.2020.107076. Epub 2020 Oct 9. Int Immunopharmacol. 2020. PMID: 33045565
-
Streptococcus uberis induced expressions of pro-inflammatory IL-6, TNF-α, and IFN-γ in bovine mammary epithelial cells associated with inhibited autophagy and autophagy flux formation.Microb Pathog. 2023 Oct;183:106270. doi: 10.1016/j.micpath.2023.106270. Epub 2023 Jul 26. Microb Pathog. 2023. PMID: 37499842
-
Role of Toll-like receptor 2 against Streptococcus uberis infection in primary mouse mammary epithelial cells.Int Immunopharmacol. 2020 Feb;79:106142. doi: 10.1016/j.intimp.2019.106142. Epub 2020 Jan 10. Int Immunopharmacol. 2020. PMID: 31931293
-
Streptococcus lutetiensis Induces Autophagy via Oxidative Stress in Bovine Mammary Epithelial Cells.Oxid Med Cell Longev. 2022 Feb 7;2022:2549772. doi: 10.1155/2022/2549772. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35178153 Free PMC article.
-
Potential factors involved in the early pathogenesis of Streptococcus uberis mastitis: a review.Folia Microbiol (Praha). 2021 Aug;66(4):509-523. doi: 10.1007/s12223-021-00879-9. Epub 2021 Jun 3. Folia Microbiol (Praha). 2021. PMID: 34085166 Review.
Cited by
-
Identifying Baicalein as a Key Bioactive Compound in XueBiJing Targeting KEAP1: Implications for Antioxidant Effects.Antioxidants (Basel). 2025 Feb 20;14(3):248. doi: 10.3390/antiox14030248. Antioxidants (Basel). 2025. PMID: 40227198 Free PMC article.
-
Danggui Buxue Decoction Alleviates Inflammation and Oxidative Stress in Mice with Escherichia coli-Induced Mastitis.Vet Sci. 2025 Mar 2;12(3):227. doi: 10.3390/vetsci12030227. Vet Sci. 2025. PMID: 40266913 Free PMC article.
-
Lachnospiraceae bacterium alleviates alcohol-associated liver disease by enhancing N-acetyl-glutamic acid levels and inhibiting ferroptosis through the KEAP1-NRF2 pathway.Gut Microbes. 2025 Dec;17(1):2517821. doi: 10.1080/19490976.2025.2517821. Epub 2025 Jun 13. Gut Microbes. 2025. PMID: 40511521 Free PMC article.
References
-
- Majumder S., Eckersall P.D., George S. Bovine mastitis: Examining factors contributing to treatment failure and prospects of nano-enabled antibacterial combination therapy. ACS Agric. Sci. Technol. 2023;7:562–582. doi: 10.1021/acsagscitech.3c00066. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

